Shaolin Gong
Deep Self-cleansing for Medical Image Segmentation with Noisy Labels
Sep 08, 2024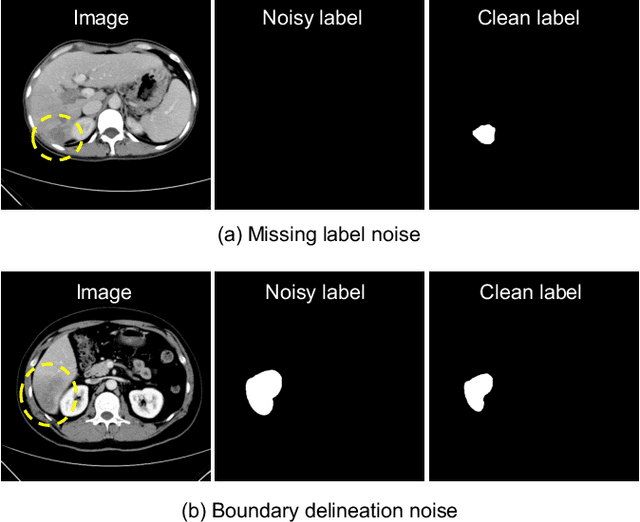
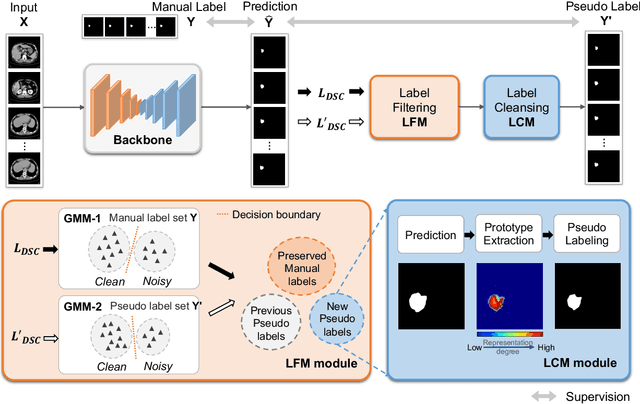
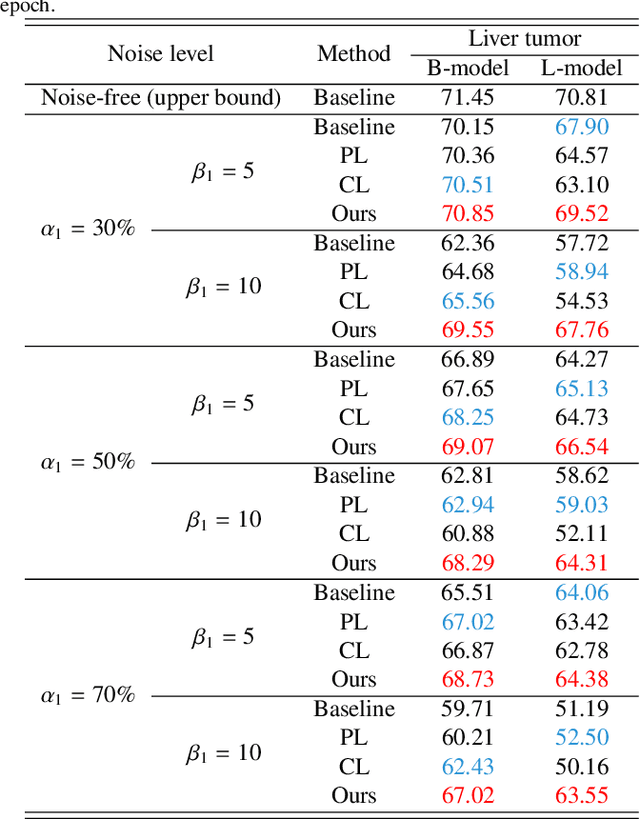
Abstract:Medical image segmentation is crucial in the field of medical imaging, aiding in disease diagnosis and surgical planning. Most established segmentation methods rely on supervised deep learning, in which clean and precise labels are essential for supervision and significantly impact the performance of models. However, manually delineated labels often contain noise, such as missing labels and inaccurate boundary delineation, which can hinder networks from correctly modeling target characteristics. In this paper, we propose a deep self-cleansing segmentation framework that can preserve clean labels while cleansing noisy ones in the training phase. To achieve this, we devise a gaussian mixture model-based label filtering module that distinguishes noisy labels from clean labels. Additionally, we develop a label cleansing module to generate pseudo low-noise labels for identified noisy samples. The preserved clean labels and pseudo-labels are then used jointly to supervise the network. Validated on a clinical liver tumor dataset and a public cardiac diagnosis dataset, our method can effectively suppress the interference from noisy labels and achieve prominent segmentation performance.
Automatic CT Segmentation from Bounding Box Annotations using Convolutional Neural Networks
Jun 09, 2021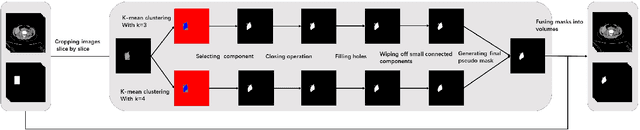
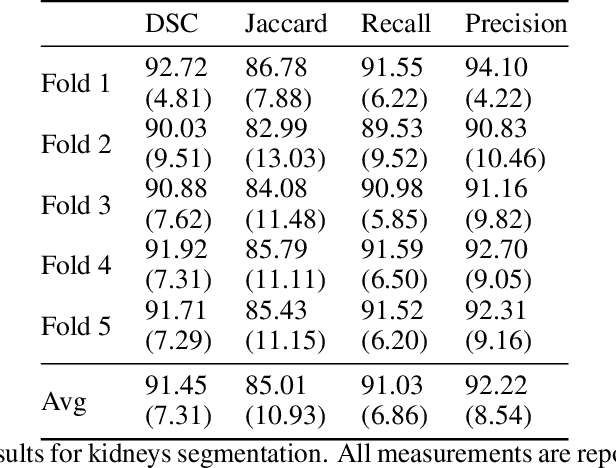
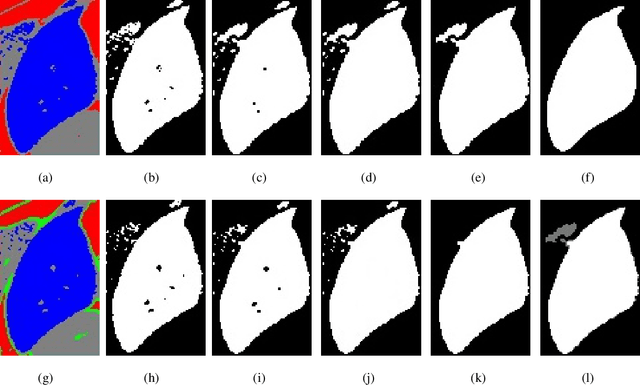
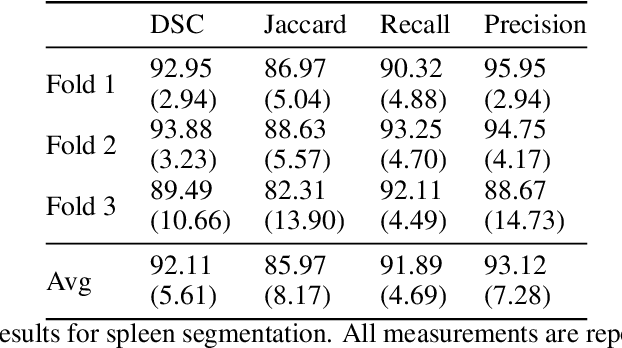
Abstract:Accurate segmentation for medical images is important for clinical diagnosis. Existing automatic segmentation methods are mainly based on fully supervised learning and have an extremely high demand for precise annotations, which are very costly and time-consuming to obtain. To address this problem, we proposed an automatic CT segmentation method based on weakly supervised learning, by which one could train an accurate segmentation model only with weak annotations in the form of bounding boxes. The proposed method is composed of two steps: 1) generating pseudo masks with bounding box annotations by k-means clustering, and 2) iteratively training a 3D U-Net convolutional neural network as a segmentation model. Some data pre-processing methods are used to improve performance. The method was validated on four datasets containing three types of organs with a total of 627 CT volumes. For liver, spleen and kidney segmentation, it achieved an accuracy of 95.19%, 92.11%, and 91.45%, respectively. Experimental results demonstrate that our method is accurate, efficient, and suitable for clinical use.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge