Dexing Kong
Haibu Mathematical-Medical Intelligent Agent:Enhancing Large Language Model Reliability in Medical Tasks via Verifiable Reasoning Chains
Oct 09, 2025Abstract:Large Language Models (LLMs) show promise in medicine but are prone to factual and logical errors, which is unacceptable in this high-stakes field. To address this, we introduce the "Haibu Mathematical-Medical Intelligent Agent" (MMIA), an LLM-driven architecture that ensures reliability through a formally verifiable reasoning process. MMIA recursively breaks down complex medical tasks into atomic, evidence-based steps. This entire reasoning chain is then automatically audited for logical coherence and evidence traceability, similar to theorem proving. A key innovation is MMIA's "bootstrapping" mode, which stores validated reasoning chains as "theorems." Subsequent tasks can then be efficiently solved using Retrieval-Augmented Generation (RAG), shifting from costly first-principles reasoning to a low-cost verification model. We validated MMIA across four healthcare administration domains, including DRG/DIP audits and medical insurance adjudication, using expert-validated benchmarks. Results showed MMIA achieved an error detection rate exceeding 98% with a false positive rate below 1%, significantly outperforming baseline LLMs. Furthermore, the RAG matching mode is projected to reduce average processing costs by approximately 85% as the knowledge base matures. In conclusion, MMIA's verifiable reasoning framework is a significant step toward creating trustworthy, transparent, and cost-effective AI systems, making LLM technology viable for critical applications in medicine.
HAIBU-ReMUD: Reasoning Multimodal Ultrasound Dataset and Model Bridging to General Specific Domains
Jun 09, 2025Abstract:Multimodal large language models (MLLMs) have shown great potential in general domains but perform poorly in some specific domains due to a lack of domain-specific data, such as image-text data or vedio-text data. In some specific domains, there is abundant graphic and textual data scattered around, but lacks standardized arrangement. In the field of medical ultrasound, there are ultrasonic diagnostic books, ultrasonic clinical guidelines, ultrasonic diagnostic reports, and so on. However, these ultrasonic materials are often saved in the forms of PDF, images, etc., and cannot be directly used for the training of MLLMs. This paper proposes a novel image-text reasoning supervised fine-tuning data generation pipeline to create specific domain quadruplets (image, question, thinking trace, and answer) from domain-specific materials. A medical ultrasound domain dataset ReMUD is established, containing over 45,000 reasoning and non-reasoning supervised fine-tuning Question Answering (QA) and Visual Question Answering (VQA) data. The ReMUD-7B model, fine-tuned on Qwen2.5-VL-7B-Instruct, outperforms general-domain MLLMs in medical ultrasound field. To facilitate research, the ReMUD dataset, data generation codebase, and ReMUD-7B parameters will be released at https://github.com/ShiDaizi/ReMUD, addressing the data shortage issue in specific domain MLLMs.
Active Contour Models Driven by Hyperbolic Mean Curvature Flow for Image Segmentation
Jun 07, 2025Abstract:Parabolic mean curvature flow-driven active contour models (PMCF-ACMs) are widely used in image segmentation, which however depend heavily on the selection of initial curve configurations. In this paper, we firstly propose several hyperbolic mean curvature flow-driven ACMs (HMCF-ACMs), which introduce tunable initial velocity fields, enabling adaptive optimization for diverse segmentation scenarios. We shall prove that HMCF-ACMs are indeed normal flows and establish the numerical equivalence between dissipative HMCF formulations and certain wave equations using the level set method with signed distance function. Building on this framework, we furthermore develop hyperbolic dual-mode regularized flow-driven ACMs (HDRF-ACMs), which utilize smooth Heaviside functions for edge-aware force modulation to suppress over-diffusion near weak boundaries. Then, we optimize a weighted fourth-order Runge-Kutta algorithm with nine-point stencil spatial discretization when solving the above-mentioned wave equations. Experiments show that both HMCF-ACMs and HDRF-ACMs could achieve more precise segmentations with superior noise resistance and numerical stability due to task-adaptive configurations of initial velocities and initial contours.
Dynamic Programming-Based Redundancy Resolution for Path Planning of Redundant Manipulators Considering Breakpoints
Nov 26, 2024Abstract:This paper proposes a redundancy resolution algorithm for a redundant manipulator based on dynamic programming. This algorithm can compute the desired joint angles at each point on a pre-planned discrete path in Cartesian space, while ensuring that the angles, velocities, and accelerations of each joint do not exceed the manipulator's constraints. We obtain the analytical solution to the inverse kinematics problem of the manipulator using a parameterization method, transforming the redundancy resolution problem into an optimization problem of determining the parameters at each path point. The constraints on joint velocity and acceleration serve as constraints for the optimization problem. Then all feasible inverse kinematic solutions for each pose under the joint angle constraints of the manipulator are obtained through parameterization methods, and the globally optimal solution to this problem is obtained through the dynamic programming algorithm. On the other hand, if a feasible joint-space path satisfying the constraints does not exist, the proposed algorithm can compute the minimum number of breakpoints required for the path and partition the path with as few breakpoints as possible to facilitate the manipulator's operation along the path. The algorithm can also determine the optimal selection of breakpoints to minimize the global cost function, rather than simply interrupting when the manipulator is unable to continue operating. The proposed algorithm is tested using a manipulator produced by a certain manufacturer, demonstrating the effectiveness of the algorithm.
Dynamic Programming-Based Offline Redundancy Resolution of Redundant Manipulators Along Prescribed Paths with Real-Time Adjustment
Nov 26, 2024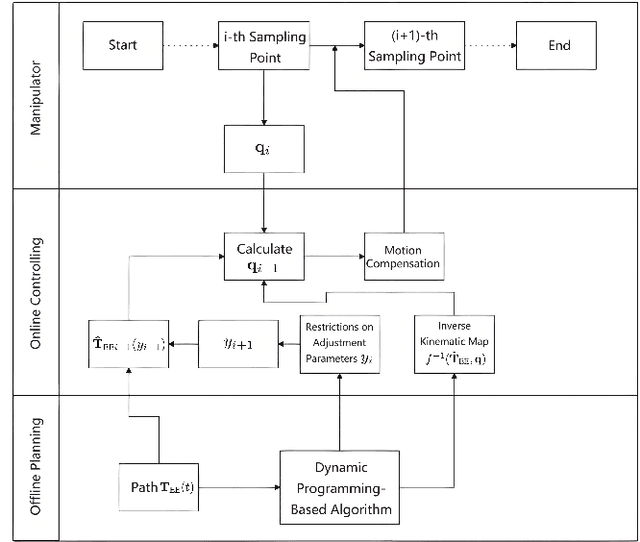
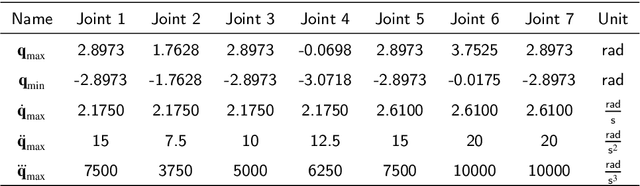
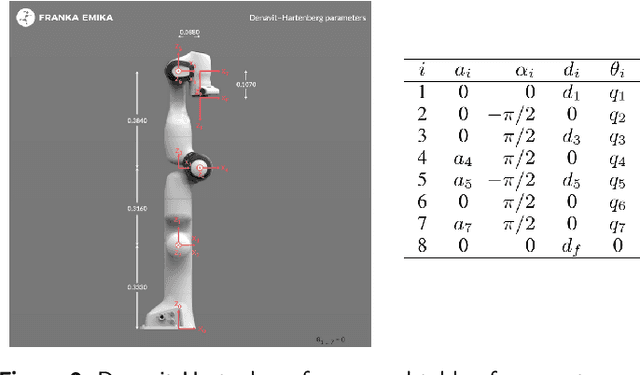
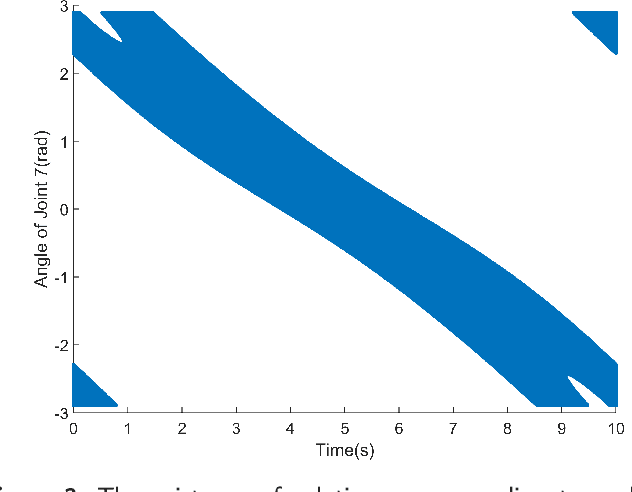
Abstract:Traditional offline redundancy resolution of trajectories for redundant manipulators involves computing inverse kinematic solutions for Cartesian space paths, constraining the manipulator to a fixed path without real-time adjustments. Online redundancy resolution can achieve real-time adjustment of paths, but it cannot consider subsequent path points, leading to the possibility of the manipulator being forced to stop mid-motion due to joint constraints. To address this, this paper introduces a dynamic programming-based offline redundancy resolution for redundant manipulators along prescribed paths with real-time adjustment. The proposed method allows the manipulator to move along a prescribed path while implementing real-time adjustment along the normal to the path. Using Dynamic Programming, the proposed approach computes a global maximum for the variation of adjustment coefficients. As long as the coefficient variation between adjacent sampling path points does not exceed this limit, the algorithm provides the next path point's joint angles based on the current joint angles, enabling the end-effector to achieve the adjusted Cartesian pose. The main innovation of this paper lies in augmenting traditional offline optimal planning with real-time adjustment capabilities, achieving a fusion of offline planning and online planning.
An edge detection-based deep learning approach for tear meniscus height measurement
Mar 23, 2024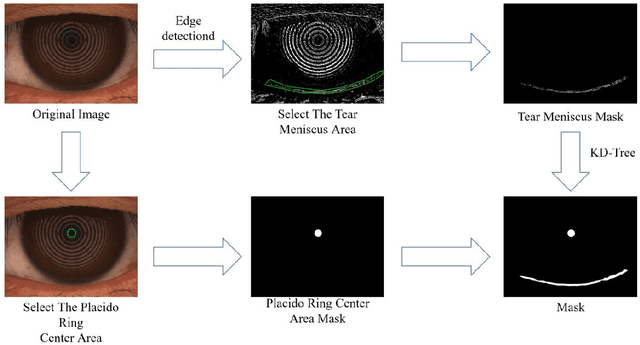
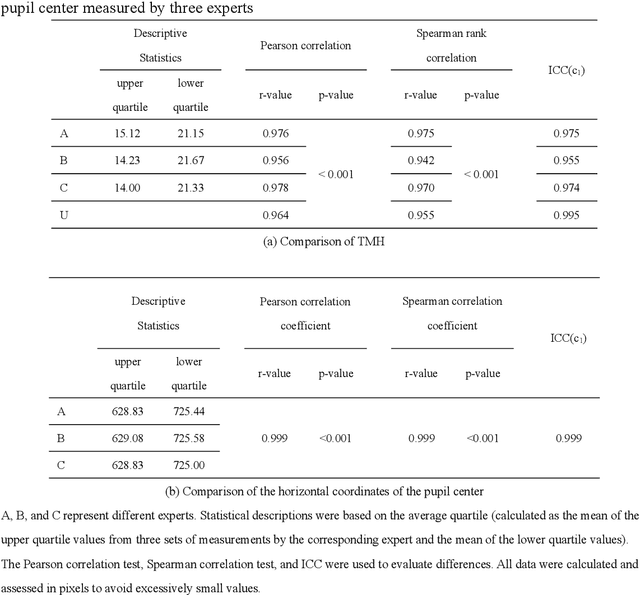
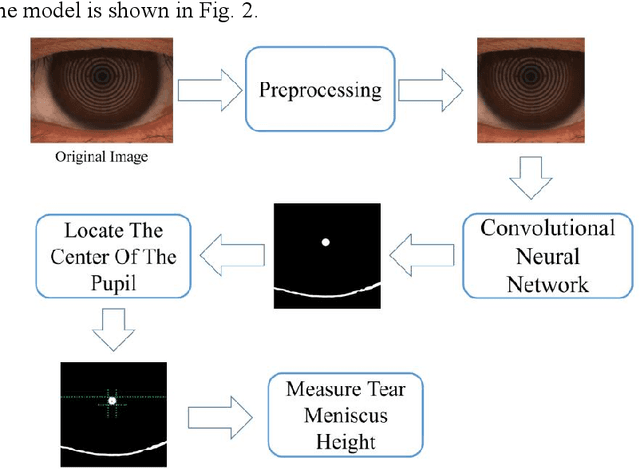

Abstract:Automatic measurements of tear meniscus height (TMH) have been achieved by using deep learning techniques; however, annotation is significantly influenced by subjective factors and is both time-consuming and labor-intensive. In this paper, we introduce an automatic TMH measurement technique based on edge detection-assisted annotation within a deep learning framework. This method generates mask labels less affected by subjective factors with enhanced efficiency compared to previous annotation approaches. For improved segmentation of the pupil and tear meniscus areas, the convolutional neural network Inceptionv3 was first implemented as an image quality assessment model, effectively identifying higher-quality images with an accuracy of 98.224%. Subsequently, by using the generated labels, various algorithms, including Unet, ResUnet, Deeplabv3+FcnResnet101, Deeplabv3+FcnResnet50, FcnResnet50, and FcnResnet101 were trained, with Unet demonstrating the best performance. Finally, Unet was used for automatic pupil and tear meniscus segmentation to locate the center of the pupil and calculate TMH,respectively. An evaluation of the mask quality predicted by Unet indicated a Mean Intersection over Union of 0.9362, a recall of 0.9261, a precision of 0.9423, and an F1-Score of 0.9326. Additionally, the TMH predicted by the model was assessed, with the fitting curve represented as y= 0.982x-0.862, an overall correlation coefficient of r^2=0.961 , and an accuracy of 94.80% (237/250). In summary, the algorithm can automatically screen images based on their quality,segment the pupil and tear meniscus areas, and automatically measure TMH. Measurement results using the AI algorithm demonstrate a high level of consistency with manual measurements, offering significant support to clinical doctors in diagnosing dry eye disease.
Automatic nodule identification and differentiation in ultrasound videos to facilitate per-nodule examination
Oct 10, 2023



Abstract:Ultrasound is a vital diagnostic technique in health screening, with the advantages of non-invasive, cost-effective, and radiation free, and therefore is widely applied in the diagnosis of nodules. However, it relies heavily on the expertise and clinical experience of the sonographer. In ultrasound images, a single nodule might present heterogeneous appearances in different cross-sectional views which makes it hard to perform per-nodule examination. Sonographers usually discriminate different nodules by examining the nodule features and the surrounding structures like gland and duct, which is cumbersome and time-consuming. To address this problem, we collected hundreds of breast ultrasound videos and built a nodule reidentification system that consists of two parts: an extractor based on the deep learning model that can extract feature vectors from the input video clips and a real-time clustering algorithm that automatically groups feature vectors by nodules. The system obtains satisfactory results and exhibits the capability to differentiate ultrasound videos. As far as we know, it's the first attempt to apply re-identification technique in the ultrasonic field.
Automatic CT Segmentation from Bounding Box Annotations using Convolutional Neural Networks
Jun 09, 2021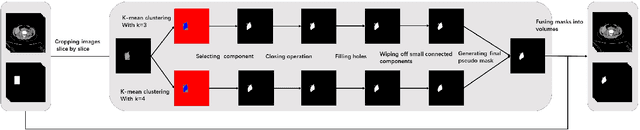
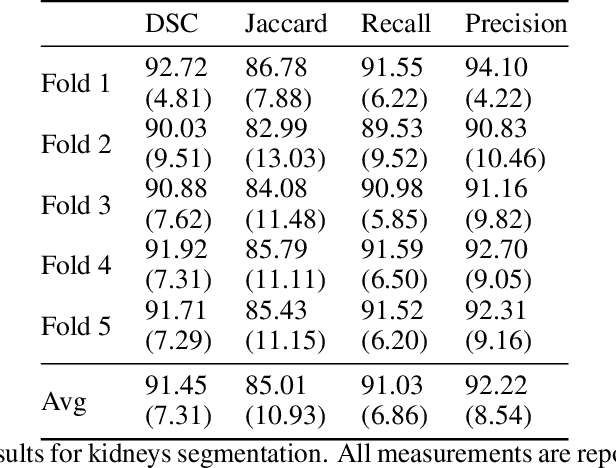
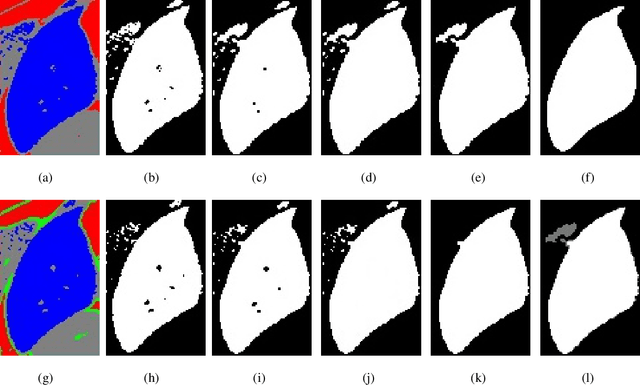
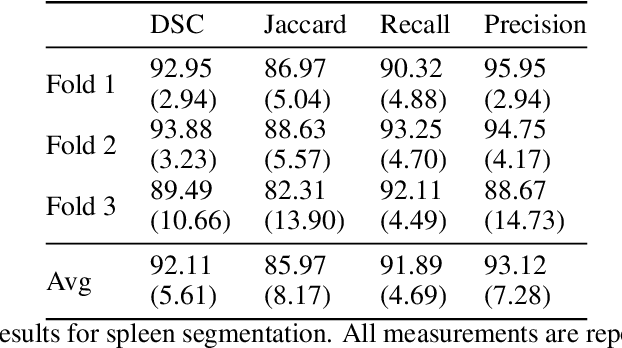
Abstract:Accurate segmentation for medical images is important for clinical diagnosis. Existing automatic segmentation methods are mainly based on fully supervised learning and have an extremely high demand for precise annotations, which are very costly and time-consuming to obtain. To address this problem, we proposed an automatic CT segmentation method based on weakly supervised learning, by which one could train an accurate segmentation model only with weak annotations in the form of bounding boxes. The proposed method is composed of two steps: 1) generating pseudo masks with bounding box annotations by k-means clustering, and 2) iteratively training a 3D U-Net convolutional neural network as a segmentation model. Some data pre-processing methods are used to improve performance. The method was validated on four datasets containing three types of organs with a total of 627 CT volumes. For liver, spleen and kidney segmentation, it achieved an accuracy of 95.19%, 92.11%, and 91.45%, respectively. Experimental results demonstrate that our method is accurate, efficient, and suitable for clinical use.
Segmentation of Levator Hiatus Using Multi-Scale Local Region Active contours and Boundary Shape Similarity Constraint
Jan 11, 2019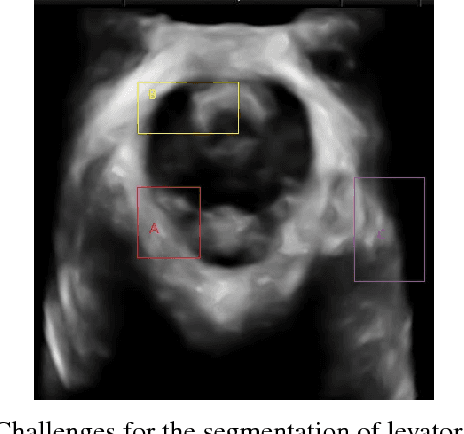
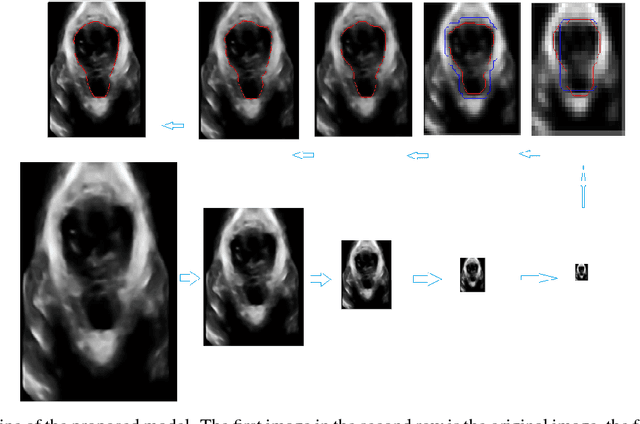
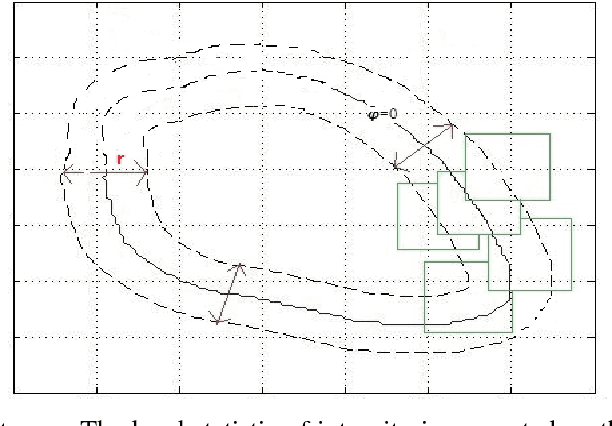
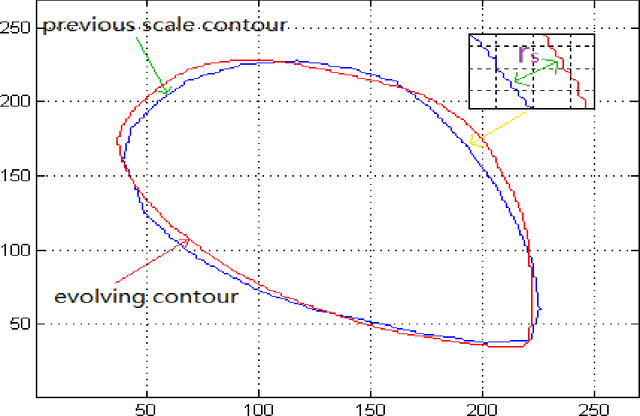
Abstract:In this paper, a multi-scale framework with local region based active contour and boundary shape similarity constraint is proposed for the segmentation of levator hiatus in ultrasound images. In this paper, we proposed a multiscale active contour framework to segment levator hiatus ultrasound images by combining the local region information and boundary shape similarity constraint. In order to get more precisely initializations and reduce the computational cost, Gaussian pyramid method is used to decompose the image into coarse-to-fine scales. A localized region active contour model is firstly performed on the coarsest scale image to get a rough contour of the levator hiatus, then the segmentation result on the coarse scale is interpolated into the finer scale image as the initialization. The boundary shape similarity between different scales is incorporate into the local region based active contour model so that the result from coarse scale can guide the contour evolution at finer scale. By incorporating the multi-scale and boundary shape similarity, the proposed method can precisely locate the levator hiatus boundaries despite various ultrasound image artifacts. With a data set of 90 levator hiatus ultrasound images, the efficiency and accuracy of the proposed method are validated by quantitative and qualitative evaluations (TP, FP, Js) and comparison with other two state-of-art active contour segmentation methods (C-V model, DRLSE model).
Automatic 3D liver location and segmentation via convolutional neural networks and graph cut
May 10, 2016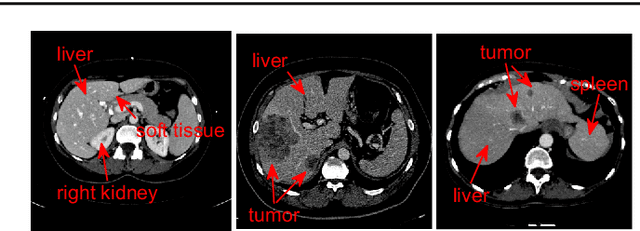
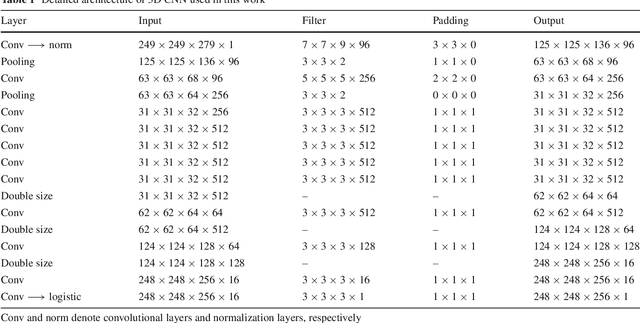
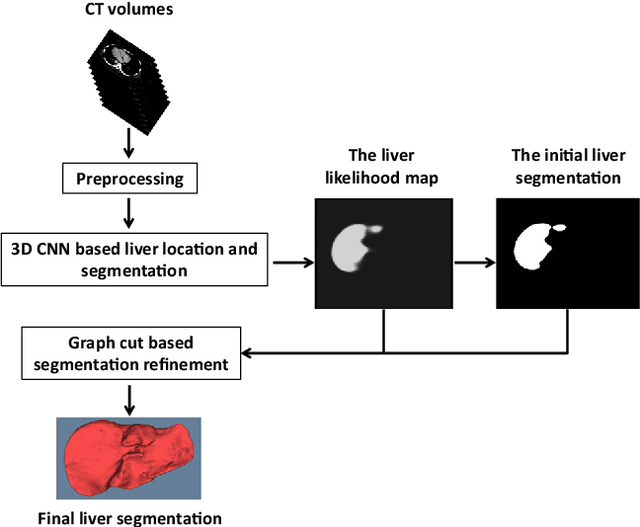
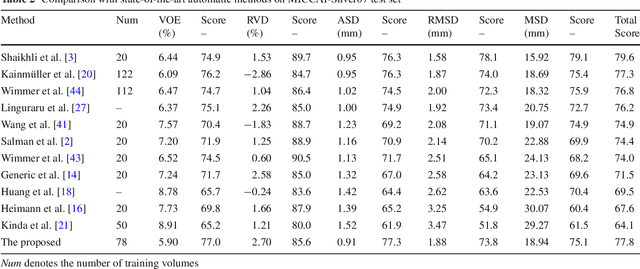
Abstract:Purpose Segmentation of the liver from abdominal computed tomography (CT) image is an essential step in some computer assisted clinical interventions, such as surgery planning for living donor liver transplant (LDLT), radiotherapy and volume measurement. In this work, we develop a deep learning algorithm with graph cut refinement to automatically segment liver in CT scans. Methods The proposed method consists of two main steps: (i) simultaneously liver detection and probabilistic segmentation using 3D convolutional neural networks (CNNs); (ii) accuracy refinement of initial segmentation with graph cut and the previously learned probability map. Results The proposed approach was validated on forty CT volumes taken from two public databases MICCAI-Sliver07 and 3Dircadb. For the MICCAI-Sliver07 test set, the calculated mean ratios of volumetric overlap error (VOE), relative volume difference (RVD), average symmetric surface distance (ASD), root mean square symmetric surface distance (RMSD) and maximum symmetric surface distance (MSD) are 5.9%, 2.7%, 0.91%, 1.88 mm, and 18.94 mm, respectively. In the case of 20 3Dircadb data, the calculated mean ratios of VOE, RVD, ASD, RMSD and MSD are 9.36%, 0.97%, 1.89%, 4.15 mm and 33.14 mm, respectively. Conclusion The proposed method is fully automatic without any user interaction. Quantitative results reveal that the proposed approach is efficient and accurate for hepatic volume estimation in a clinical setup. The high correlation between the automatic and manual references shows that the proposed method can be good enough to replace the time-consuming and non-reproducible manual segmentation method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge