Fang Lu
IBM Research
A two-stream network with global-local feature fusion for bone age assessment
Dec 20, 2025



Abstract:Bone Age Assessment (BAA) is a widely used clinical technique that can accurately reflect an individual's growth and development level, as well as maturity. In recent years, although deep learning has advanced the field of bone age assessment, existing methods face challenges in efficiently balancing global features and local skeletal details. This study aims to develop an automated bone age assessment system based on a two-stream deep learning architecture to achieve higher accuracy in bone age assessment. We propose the BoNet+ model incorporating global and local feature extraction channels. A Transformer module is introduced into the global feature extraction channel to enhance the ability in extracting global features through multi-head self-attention mechanism. A RFAConv module is incorporated into the local feature extraction channel to generate adaptive attention maps within multiscale receptive fields, enhancing local feature extraction capabilities. Global and local features are concatenated along the channel dimension and optimized by an Inception-V3 network. The proposed method has been validated on the Radiological Society of North America (RSNA) and Radiological Hand Pose Estimation (RHPE) test datasets, achieving mean absolute errors (MAEs) of 3.81 and 5.65 months, respectively. These results are comparable to the state-of-the-art. The BoNet+ model reduces the clinical workload and achieves automatic, high-precision, and more objective bone age assessment.
FMD-TransUNet: Abdominal Multi-Organ Segmentation Based on Frequency Domain Multi-Axis Representation Learning and Dual Attention Mechanisms
Sep 19, 2025Abstract:Accurate abdominal multi-organ segmentation is critical for clinical applications. Although numerous deep learning-based automatic segmentation methods have been developed, they still struggle to segment small, irregular, or anatomically complex organs. Moreover, most current methods focus on spatial-domain analysis, often overlooking the synergistic potential of frequency-domain representations. To address these limitations, we propose a novel framework named FMD-TransUNet for precise abdominal multi-organ segmentation. It innovatively integrates the Multi-axis External Weight Block (MEWB) and the improved dual attention module (DA+) into the TransUNet framework. The MEWB extracts multi-axis frequency-domain features to capture both global anatomical structures and local boundary details, providing complementary information to spatial-domain representations. The DA+ block utilizes depthwise separable convolutions and incorporates spatial and channel attention mechanisms to enhance feature fusion, reduce redundant information, and narrow the semantic gap between the encoder and decoder. Experimental validation on the Synapse dataset shows that FMD-TransUNet outperforms other recent state-of-the-art methods, achieving an average DSC of 81.32\% and a HD of 16.35 mm across eight abdominal organs. Compared to the baseline model, the average DSC increased by 3.84\%, and the average HD decreased by 15.34 mm. These results demonstrate the effectiveness of FMD-TransUNet in improving the accuracy of abdominal multi-organ segmentation.
Effect of secular trend in drug effectiveness study in real world data
Aug 18, 2018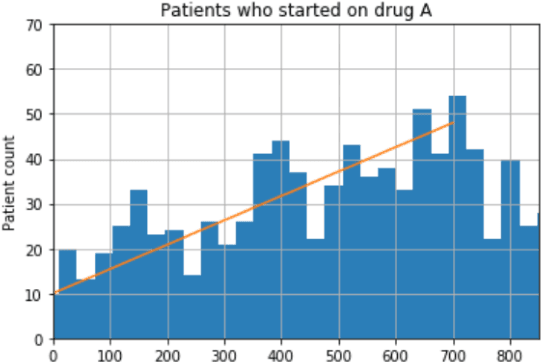
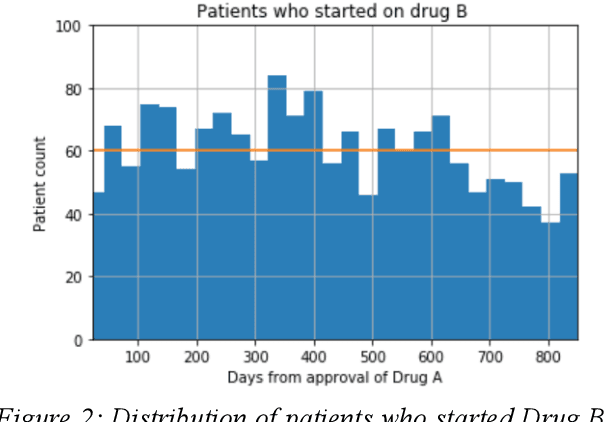

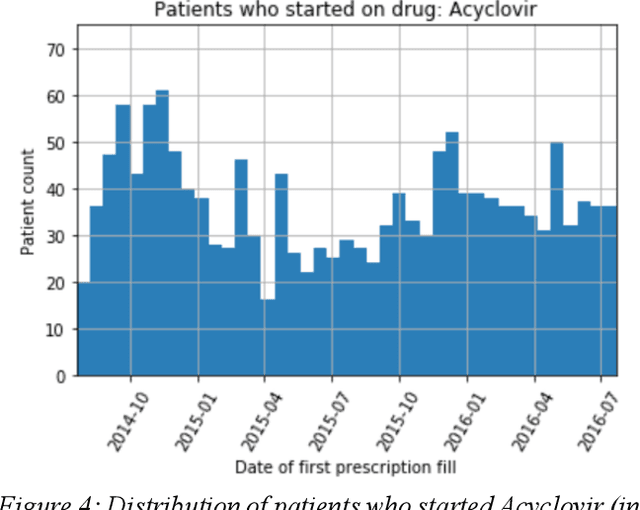
Abstract:We discovered secular trend bias in a drug effectiveness study for a recently approved drug. We compared treatment outcomes between patients who received the newly approved drug and patients exposed to the standard treatment. All patients diagnosed after the new drug's approval date were considered. We built a machine learning causal inference model to determine patient subpopulations likely to respond better to the newly approved drug. After identifying the presence of secular trend bias in our data, we attempted to adjust for the bias in two different ways. First, we matched patients on the number of days from the new drug's approval date that the patient's treatment (new or standard) began. Second, we included a covariate in the model for the number of days between the date of approval of the new drug and the treatment (new or standard) start date. Neither approach completely mitigated the bias. Residual bias we attribute to differences in patient disease severity or other unmeasured patient characteristics. Had we not identified the secular trend bias in our data, the causal inference model would have been interpreted without consideration for this underlying bias. Being aware of, testing for, and handling potential bias in the data is essential to diminish the uncertainty in AI modeling.
Automatic 3D liver location and segmentation via convolutional neural networks and graph cut
May 10, 2016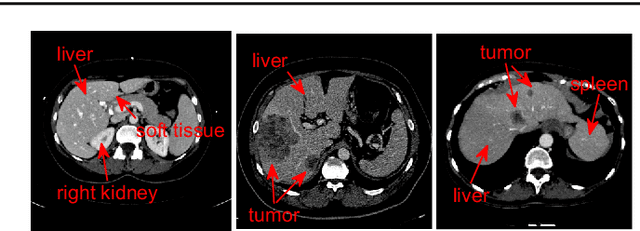
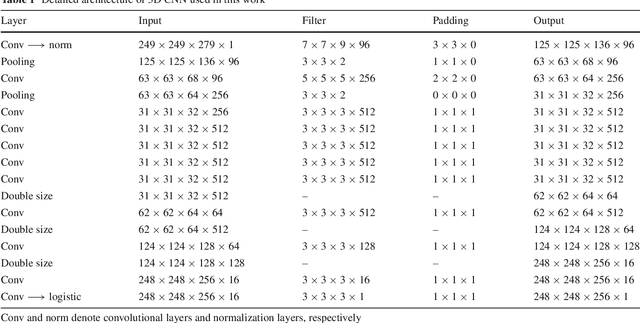
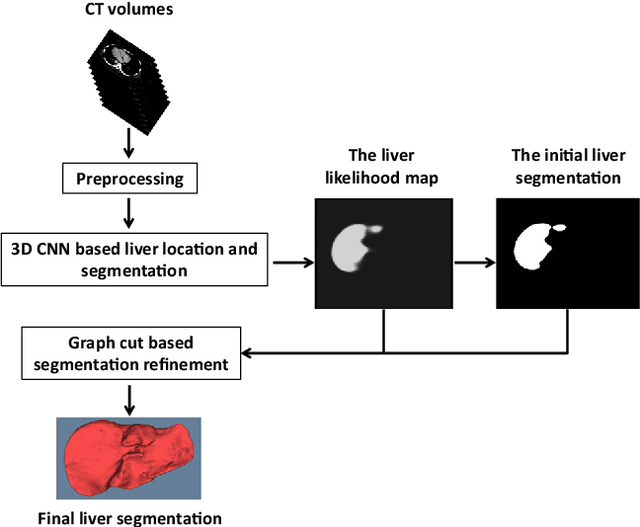
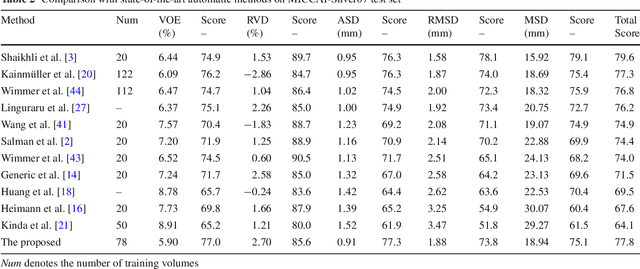
Abstract:Purpose Segmentation of the liver from abdominal computed tomography (CT) image is an essential step in some computer assisted clinical interventions, such as surgery planning for living donor liver transplant (LDLT), radiotherapy and volume measurement. In this work, we develop a deep learning algorithm with graph cut refinement to automatically segment liver in CT scans. Methods The proposed method consists of two main steps: (i) simultaneously liver detection and probabilistic segmentation using 3D convolutional neural networks (CNNs); (ii) accuracy refinement of initial segmentation with graph cut and the previously learned probability map. Results The proposed approach was validated on forty CT volumes taken from two public databases MICCAI-Sliver07 and 3Dircadb. For the MICCAI-Sliver07 test set, the calculated mean ratios of volumetric overlap error (VOE), relative volume difference (RVD), average symmetric surface distance (ASD), root mean square symmetric surface distance (RMSD) and maximum symmetric surface distance (MSD) are 5.9%, 2.7%, 0.91%, 1.88 mm, and 18.94 mm, respectively. In the case of 20 3Dircadb data, the calculated mean ratios of VOE, RVD, ASD, RMSD and MSD are 9.36%, 0.97%, 1.89%, 4.15 mm and 33.14 mm, respectively. Conclusion The proposed method is fully automatic without any user interaction. Quantitative results reveal that the proposed approach is efficient and accurate for hepatic volume estimation in a clinical setup. The high correlation between the automatic and manual references shows that the proposed method can be good enough to replace the time-consuming and non-reproducible manual segmentation method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge