Daniel King
LoRA Learns Less and Forgets Less
May 15, 2024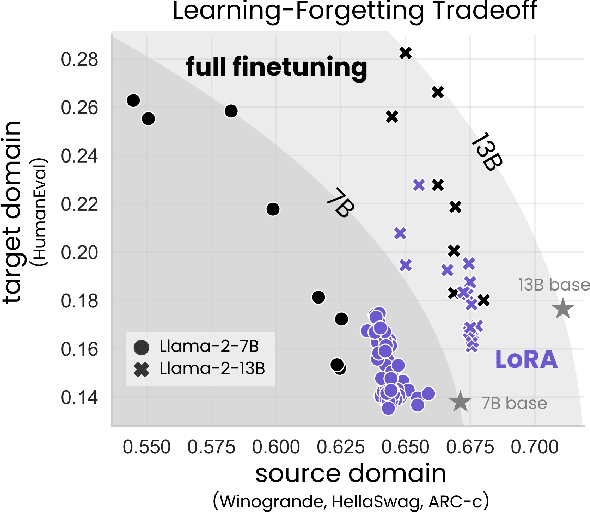

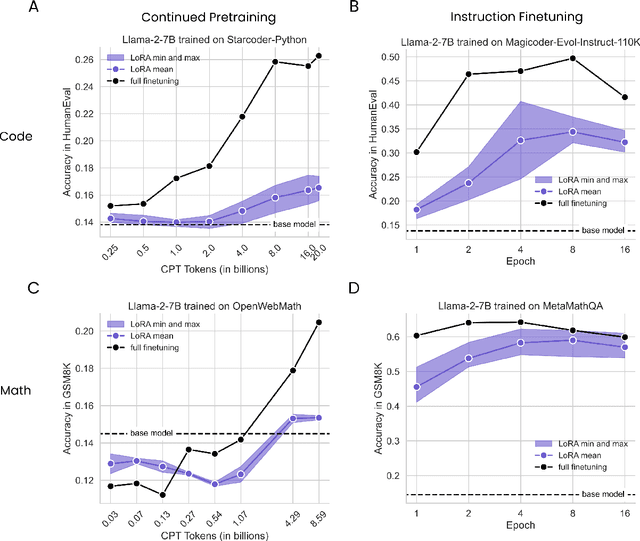

Abstract:Low-Rank Adaptation (LoRA) is a widely-used parameter-efficient finetuning method for large language models. LoRA saves memory by training only low rank perturbations to selected weight matrices. In this work, we compare the performance of LoRA and full finetuning on two target domains, programming and mathematics. We consider both the instruction finetuning ($\approx$100K prompt-response pairs) and continued pretraining ($\approx$10B unstructured tokens) data regimes. Our results show that, in most settings, LoRA substantially underperforms full finetuning. Nevertheless, LoRA exhibits a desirable form of regularization: it better maintains the base model's performance on tasks outside the target domain. We show that LoRA provides stronger regularization compared to common techniques such as weight decay and dropout; it also helps maintain more diverse generations. We show that full finetuning learns perturbations with a rank that is 10-100X greater than typical LoRA configurations, possibly explaining some of the reported gaps. We conclude by proposing best practices for finetuning with LoRA.
MosaicBERT: A Bidirectional Encoder Optimized for Fast Pretraining
Jan 16, 2024



Abstract:Although BERT-style encoder models are heavily used in NLP research, many researchers do not pretrain their own BERTs from scratch due to the high cost of training. In the past half-decade since BERT first rose to prominence, many advances have been made with other transformer architectures and training configurations that have yet to be systematically incorporated into BERT. Here, we introduce MosaicBERT, a BERT-style encoder architecture and training recipe that is empirically optimized for fast pretraining. This efficient architecture incorporates FlashAttention, Attention with Linear Biases (ALiBi), Gated Linear Units (GLU), a module to dynamically remove padded tokens, and low precision LayerNorm into the classic transformer encoder block. The training recipe includes a 30% masking ratio for the Masked Language Modeling (MLM) objective, bfloat16 precision, and vocabulary size optimized for GPU throughput, in addition to best-practices from RoBERTa and other encoder models. When pretrained from scratch on the C4 dataset, this base model achieves a downstream average GLUE (dev) score of 79.6 in 1.13 hours on 8 A100 80 GB GPUs at a cost of roughly $20. We plot extensive accuracy vs. pretraining speed Pareto curves and show that MosaicBERT base and large are consistently Pareto optimal when compared to a competitive BERT base and large. This empirical speed up in pretraining enables researchers and engineers to pretrain custom BERT-style models at low cost instead of finetune on existing generic models. We open source our model weights and code.
* 10 pages, 4 figures in main text. 25 pages total
The Semantic Scholar Open Data Platform
Jan 24, 2023



Abstract:The volume of scientific output is creating an urgent need for automated tools to help scientists keep up with developments in their field. Semantic Scholar (S2) is an open data platform and website aimed at accelerating science by helping scholars discover and understand scientific literature. We combine public and proprietary data sources using state-of-the-art techniques for scholarly PDF content extraction and automatic knowledge graph construction to build the Semantic Scholar Academic Graph, the largest open scientific literature graph to-date, with 200M+ papers, 80M+ authors, 550M+ paper-authorship edges, and 2.4B+ citation edges. The graph includes advanced semantic features such as structurally parsed text, natural language summaries, and vector embeddings. In this paper, we describe the components of the S2 data processing pipeline and the associated APIs offered by the platform. We will update this living document to reflect changes as we add new data offerings and improve existing services.
ACCoRD: A Multi-Document Approach to Generating Diverse Descriptions of Scientific Concepts
May 14, 2022
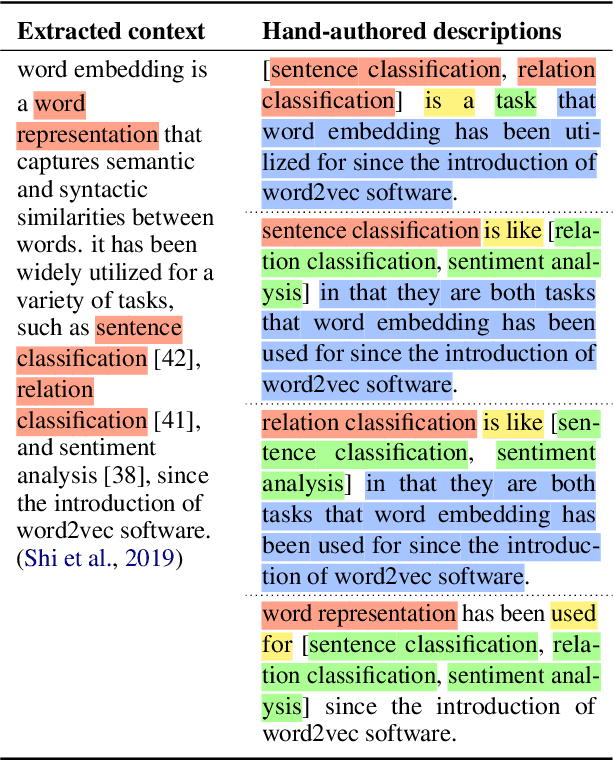

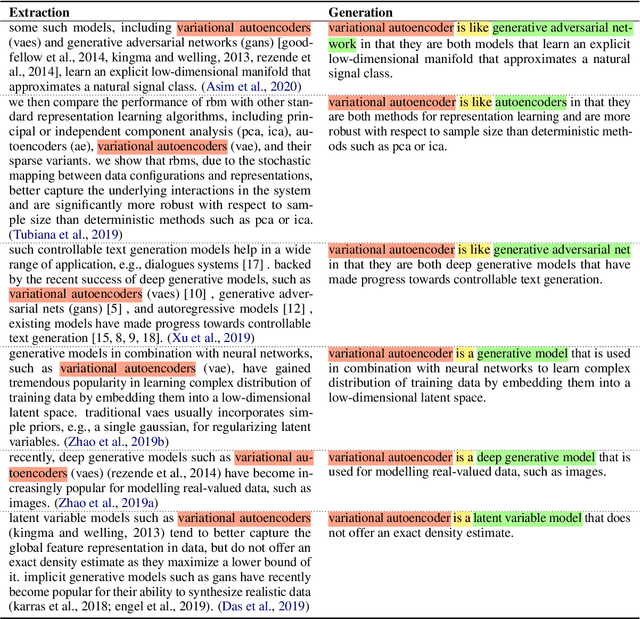
Abstract:Systems that can automatically define unfamiliar terms hold the promise of improving the accessibility of scientific texts, especially for readers who may lack prerequisite background knowledge. However, current systems assume a single "best" description per concept, which fails to account for the many potentially useful ways a concept can be described. We present ACCoRD, an end-to-end system tackling the novel task of generating sets of descriptions of scientific concepts. Our system takes advantage of the myriad ways a concept is mentioned across the scientific literature to produce distinct, diverse descriptions of target scientific concepts in terms of different reference concepts. To support research on the task, we release an expert-annotated resource, the ACCoRD corpus, which includes 1,275 labeled contexts and 1,787 hand-authored concept descriptions. We conduct a user study demonstrating that (1) users prefer descriptions produced by our end-to-end system, and (2) users prefer multiple descriptions to a single "best" description.
Don't Say What You Don't Know: Improving the Consistency of Abstractive Summarization by Constraining Beam Search
Mar 16, 2022



Abstract:Abstractive summarization systems today produce fluent and relevant output, but often "hallucinate" statements not supported by the source text. We analyze the connection between hallucinations and training data, and find evidence that models hallucinate because they train on target summaries that are unsupported by the source. Based on our findings, we present PINOCCHIO, a new decoding method that improves the consistency of a transformer-based abstractive summarizer by constraining beam search to avoid hallucinations. Given the model states and outputs at a given step, PINOCCHIO detects likely model hallucinations based on various measures of attribution to the source text. PINOCCHIO backtracks to find more consistent output, and can opt to produce no summary at all when no consistent generation can be found. In experiments, we find that PINOCCHIO improves the consistency of generation (in terms of F1) by an average of~67% on two abstractive summarization datasets.
Reducing Annotating Load: Active Learning with Synthetic Images in Surgical Instrument Segmentation
Aug 07, 2021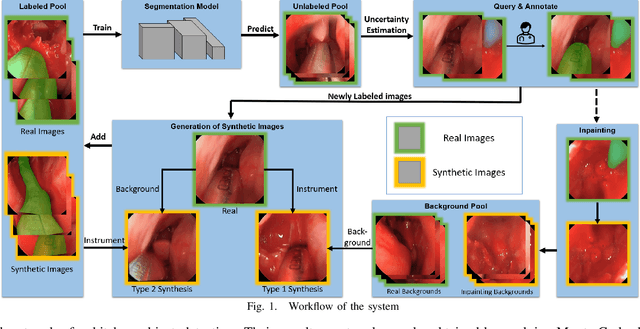
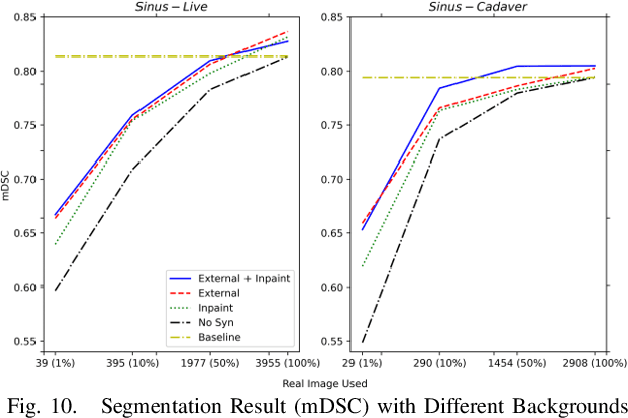
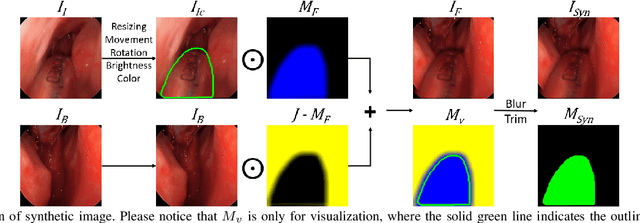
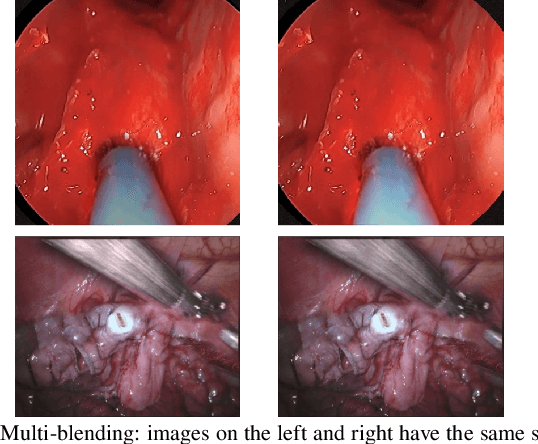
Abstract:Accurate instrument segmentation in endoscopic vision of robot-assisted surgery is challenging due to reflection on the instruments and frequent contacts with tissue. Deep neural networks (DNN) show competitive performance and are in favor in recent years. However, the hunger of DNN for labeled data poses a huge workload of annotation. Motivated by alleviating this workload, we propose a general embeddable method to decrease the usage of labeled real images, using active generated synthetic images. In each active learning iteration, the most informative unlabeled images are first queried by active learning and then labeled. Next, synthetic images are generated based on these selected images. The instruments and backgrounds are cropped out and randomly combined with each other with blending and fusion near the boundary. The effectiveness of the proposed method is validated on 2 sinus surgery datasets and 1 intraabdominal surgery dataset. The results indicate a considerable improvement in performance, especially when the budget for annotation is small. The effectiveness of different types of synthetic images, blending methods, and external background are also studied. All the code is open-sourced at: https://github.com/HaonanPeng/active_syn_generator.
High-Precision Extraction of Emerging Concepts from Scientific Literature
Jun 11, 2020


Abstract:Identification of new concepts in scientific literature can help power faceted search, scientific trend analysis, knowledge-base construction, and more, but current methods are lacking. Manual identification cannot keep up with the torrent of new publications, while the precision of existing automatic techniques is too low for many applications. We present an unsupervised concept extraction method for scientific literature that achieves much higher precision than previous work. Our approach relies on a simple but novel intuition: each scientific concept is likely to be introduced or popularized by a single paper that is disproportionately cited by subsequent papers mentioning the concept. From a corpus of computer science papers on arXiv, we find that our method achieves a Precision@1000 of 99%, compared to 86% for prior work, and a substantially better precision-yield trade-off across the top 15,000 extractions. To stimulate research in this area, we release our code and data (https://github.com/allenai/ForeCite).
Pretrained Language Models for Sequential Sentence Classification
Sep 22, 2019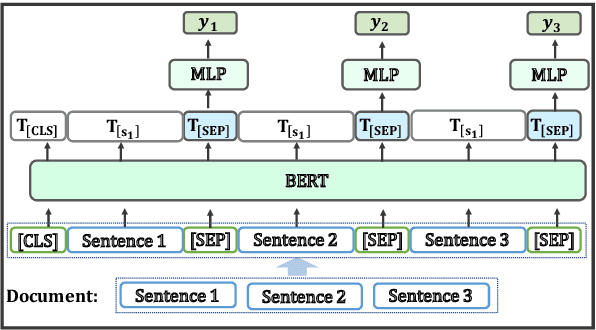

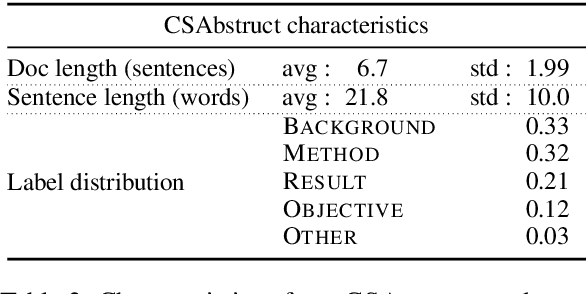
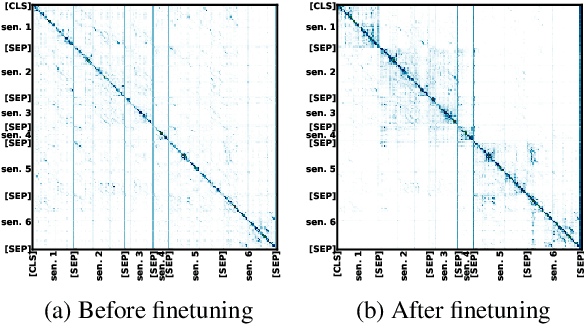
Abstract:As a step toward better document-level understanding, we explore classification of a sequence of sentences into their corresponding categories, a task that requires understanding sentences in context of the document. Recent successful models for this task have used hierarchical models to contextualize sentence representations, and Conditional Random Fields (CRFs) to incorporate dependencies between subsequent labels. In this work, we show that pretrained language models, BERT (Devlin et al., 2018) in particular, can be used for this task to capture contextual dependencies without the need for hierarchical encoding nor a CRF. Specifically, we construct a joint sentence representation that allows BERT Transformer layers to directly utilize contextual information from all words in all sentences. Our approach achieves state-of-the-art results on four datasets, including a new dataset of structured scientific abstracts.
Strong Baselines for Complex Word Identification across Multiple Languages
Apr 11, 2019
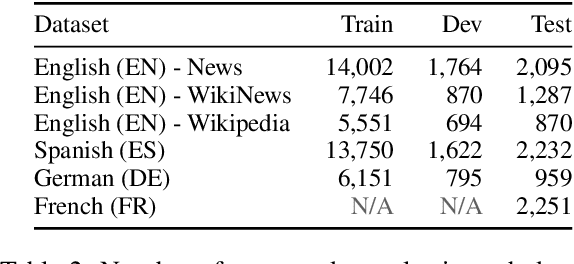
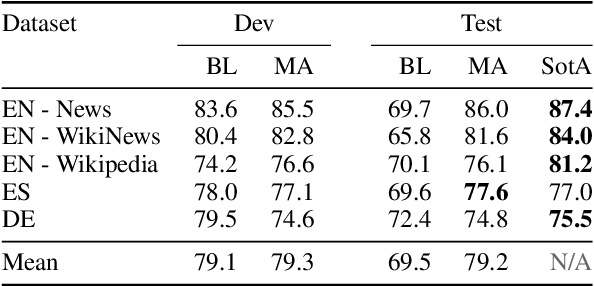

Abstract:Complex Word Identification (CWI) is the task of identifying which words or phrases in a sentence are difficult to understand by a target audience. The latest CWI Shared Task released data for two settings: monolingual (i.e. train and test in the same language) and cross-lingual (i.e. test in a language not seen during training). The best monolingual models relied on language-dependent features, which do not generalise in the cross-lingual setting, while the best cross-lingual model used neural networks with multi-task learning. In this paper, we present monolingual and cross-lingual CWI models that perform as well as (or better than) most models submitted to the latest CWI Shared Task. We show that carefully selected features and simple learning models can achieve state-of-the-art performance, and result in strong baselines for future development in this area. Finally, we discuss how inconsistencies in the annotation of the data can explain some of the results obtained.
ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing
Feb 21, 2019



Abstract:Despite recent advances in natural language processing, many statistical models for processing text perform extremely poorly under domain shift. Processing biomedical and clinical text is a critically important application area of natural language processing, for which there are few robust, practical, publicly available models. This paper describes scispaCy, a new tool for practical biomedical/scientific text processing, which heavily leverages the spaCy library. We detail the performance of two packages of models released in scispaCy and demonstrate their robustness on several tasks and datasets. Models and code are available at https://allenai.github.io/scispacy/
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge