Waleed Ammar
TyDi QA-WANA: A Benchmark for Information-Seeking Question Answering in Languages of West Asia and North Africa
Jul 23, 2025Abstract:We present TyDi QA-WANA, a question-answering dataset consisting of 28K examples divided among 10 language varieties of western Asia and northern Africa. The data collection process was designed to elicit information-seeking questions, where the asker is genuinely curious to know the answer. Each question in paired with an entire article that may or may not contain the answer; the relatively large size of the articles results in a task suitable for evaluating models' abilities to utilize large text contexts in answering questions. Furthermore, the data was collected directly in each language variety, without the use of translation, in order to avoid issues of cultural relevance. We present performance of two baseline models, and release our code and data to facilitate further improvement by the research community.
PRESTO: A Multilingual Dataset for Parsing Realistic Task-Oriented Dialogs
Mar 17, 2023



Abstract:Research interest in task-oriented dialogs has increased as systems such as Google Assistant, Alexa and Siri have become ubiquitous in everyday life. However, the impact of academic research in this area has been limited by the lack of datasets that realistically capture the wide array of user pain points. To enable research on some of the more challenging aspects of parsing realistic conversations, we introduce PRESTO, a public dataset of over 550K contextual multilingual conversations between humans and virtual assistants. PRESTO contains a diverse array of challenges that occur in real-world NLU tasks such as disfluencies, code-switching, and revisions. It is the only large scale human generated conversational parsing dataset that provides structured context such as a user's contacts and lists for each example. Our mT5 model based baselines demonstrate that the conversational phenomenon present in PRESTO are challenging to model, which is further pronounced in a low-resource setup.
Extracting evidence of supplement-drug interactions from literature
Sep 17, 2019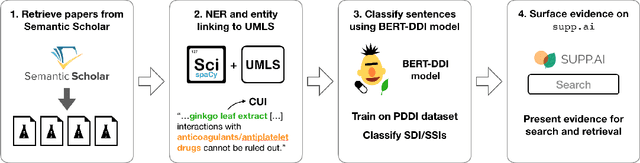
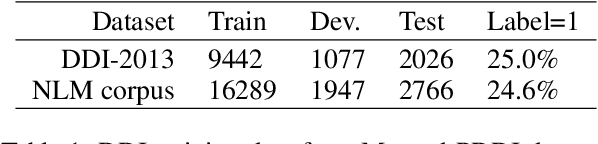
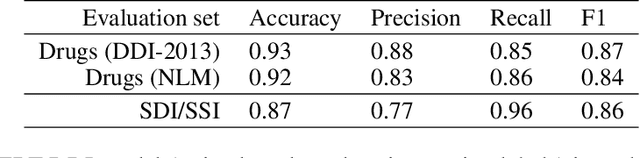
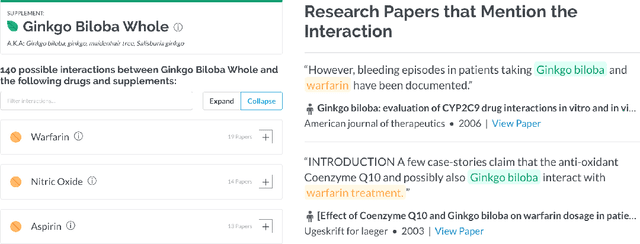
Abstract:Dietary supplements are used by a large portion of the population, but information on their safety is hard to find. We demonstrate an automated method for extracting evidence of supplement-drug interactions (SDIs) and supplement-supplement interactions (SSIs) from scientific text. To address the lack of labeled data in this domain, we use labels of the closely related task of identifying drug-drug interactions (DDIs) for supervision, and assess the feasibility of transferring the model to identify supplement interactions. We fine-tune the contextualized word representations of BERT-large using labeled data from the PDDI corpus. We then process 22M abstracts from PubMed using this model, and extract evidence for 55946 unique interactions between 1923 supplements and 2727 drugs (precision: 0.77, recall: 0.96), demonstrating that learning the task of DDI classification transfers successfully to the related problem of identifying SDIs and SSIs. As far as we know, this is the first published work on detecting evidence of SDIs/SSIs from literature. We implement a freely-available public interface supp.ai to browse and search evidence sentences extracted by our model.
Structural Scaffolds for Citation Intent Classification in Scientific Publications
Apr 02, 2019



Abstract:Identifying the intent of a citation in scientific papers (e.g., background information, use of methods, comparing results) is critical for machine reading of individual publications and automated analysis of the scientific literature. We propose structural scaffolds, a multitask model to incorporate structural information of scientific papers into citations for effective classification of citation intents. Our model achieves a new state-of-the-art on an existing ACL anthology dataset (ACL-ARC) with a 13.3% absolute increase in F1 score, without relying on external linguistic resources or hand-engineered features as done in existing methods. In addition, we introduce a new dataset of citation intents (SciCite) which is more than five times larger and covers multiple scientific domains compared with existing datasets. Our code and data are available at: https://github.com/allenai/scicite.
ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing
Feb 21, 2019



Abstract:Despite recent advances in natural language processing, many statistical models for processing text perform extremely poorly under domain shift. Processing biomedical and clinical text is a critically important application area of natural language processing, for which there are few robust, practical, publicly available models. This paper describes scispaCy, a new tool for practical biomedical/scientific text processing, which heavily leverages the spaCy library. We detail the performance of two packages of models released in scispaCy and demonstrate their robustness on several tasks and datasets. Models and code are available at https://allenai.github.io/scispacy/
Improving Distant Supervision with Maxpooled Attention and Sentence-Level Supervision
Oct 30, 2018



Abstract:We propose an effective multitask learning setup for reducing distant supervision noise by leveraging sentence-level supervision. We show how sentence-level supervision can be used to improve the encoding of individual sentences, and to learn which input sentences are more likely to express the relationship between a pair of entities. We also introduce a novel neural architecture for collecting signals from multiple input sentences, which combines the benefits of attention and maxpooling. The proposed method increases AUC by 10% (from 0.261 to 0.284), and outperforms recently published results on the FB-NYT dataset.
Ontology Alignment in the Biomedical Domain Using Entity Definitions and Context
Jun 20, 2018



Abstract:Ontology alignment is the task of identifying semantically equivalent entities from two given ontologies. Different ontologies have different representations of the same entity, resulting in a need to de-duplicate entities when merging ontologies. We propose a method for enriching entities in an ontology with external definition and context information, and use this additional information for ontology alignment. We develop a neural architecture capable of encoding the additional information when available, and show that the addition of external data results in an F1-score of 0.69 on the Ontology Alignment Evaluation Initiative (OAEI) largebio SNOMED-NCI subtask, comparable with the entity-level matchers in a SOTA system.
Extracting Scientific Figures with Distantly Supervised Neural Networks
May 30, 2018

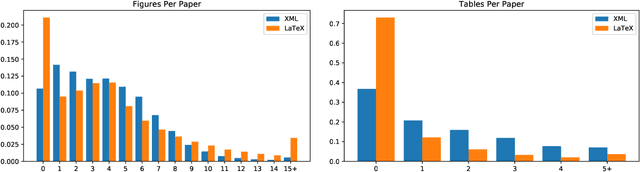

Abstract:Non-textual components such as charts, diagrams and tables provide key information in many scientific documents, but the lack of large labeled datasets has impeded the development of data-driven methods for scientific figure extraction. In this paper, we induce high-quality training labels for the task of figure extraction in a large number of scientific documents, with no human intervention. To accomplish this we leverage the auxiliary data provided in two large web collections of scientific documents (arXiv and PubMed) to locate figures and their associated captions in the rasterized PDF. We share the resulting dataset of over 5.5 million induced labels---4,000 times larger than the previous largest figure extraction dataset---with an average precision of 96.8%, to enable the development of modern data-driven methods for this task. We use this dataset to train a deep neural network for end-to-end figure detection, yielding a model that can be more easily extended to new domains compared to previous work. The model was successfully deployed in Semantic Scholar, a large-scale academic search engine, and used to extract figures in 13 million scientific documents.
Construction of the Literature Graph in Semantic Scholar
May 06, 2018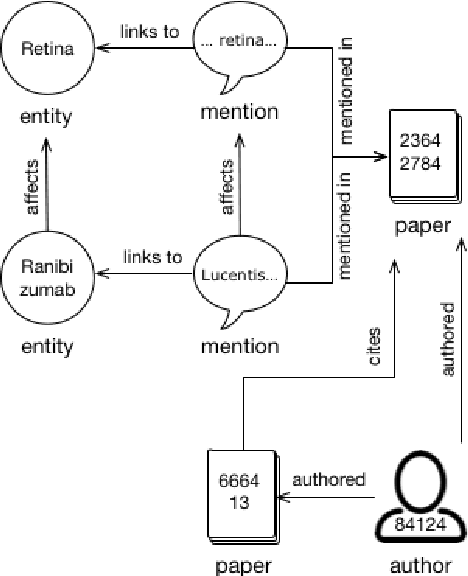
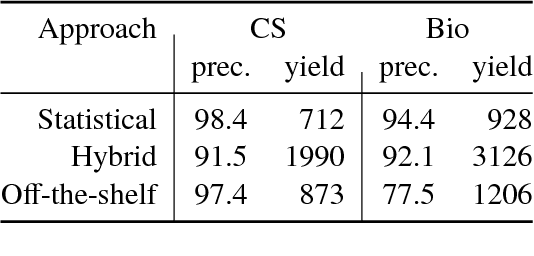
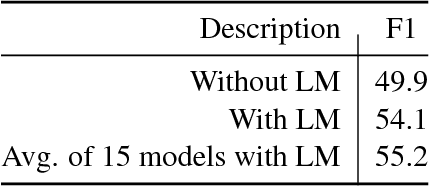
Abstract:We describe a deployed scalable system for organizing published scientific literature into a heterogeneous graph to facilitate algorithmic manipulation and discovery. The resulting literature graph consists of more than 280M nodes, representing papers, authors, entities and various interactions between them (e.g., authorships, citations, entity mentions). We reduce literature graph construction into familiar NLP tasks (e.g., entity extraction and linking), point out research challenges due to differences from standard formulations of these tasks, and report empirical results for each task. The methods described in this paper are used to enable semantic features in www.semanticscholar.org
A Dataset of Peer Reviews : Collection, Insights and NLP Applications
Apr 25, 2018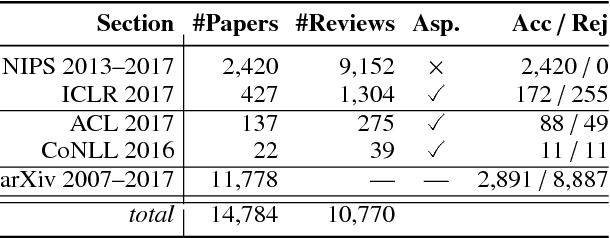
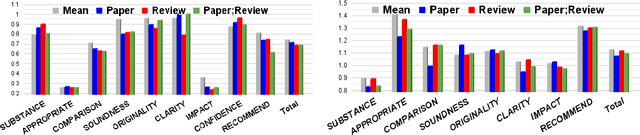
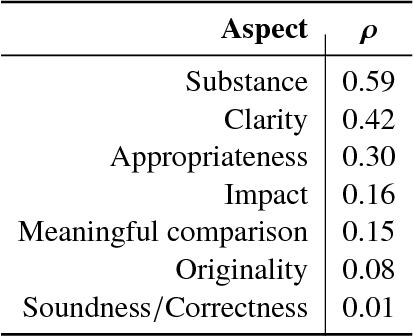
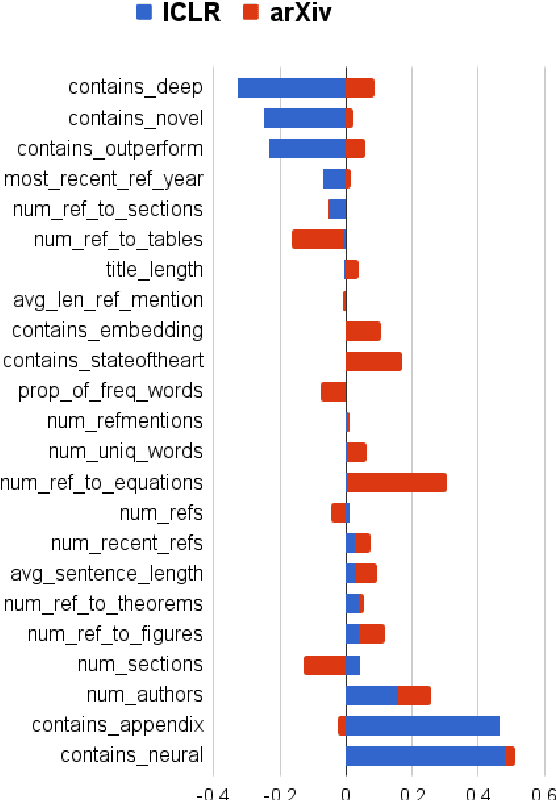
Abstract:Peer reviewing is a central component in the scientific publishing process. We present the first public dataset of scientific peer reviews available for research purposes (PeerRead v1) providing an opportunity to study this important artifact. The dataset consists of 14.7K paper drafts and the corresponding accept/reject decisions in top-tier venues including ACL, NIPS and ICLR. The dataset also includes 10.7K textual peer reviews written by experts for a subset of the papers. We describe the data collection process and report interesting observed phenomena in the peer reviews. We also propose two novel NLP tasks based on this dataset and provide simple baseline models. In the first task, we show that simple models can predict whether a paper is accepted with up to 21% error reduction compared to the majority baseline. In the second task, we predict the numerical scores of review aspects and show that simple models can outperform the mean baseline for aspects with high variance such as 'originality' and 'impact'.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge