Chris Wilhelm
TÜLU 3: Pushing Frontiers in Open Language Model Post-Training
Nov 22, 2024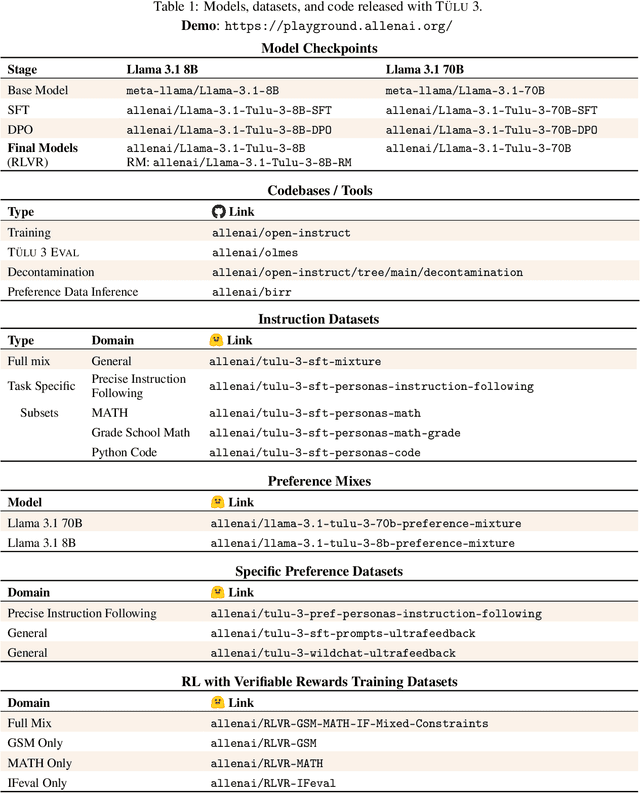



Abstract:Language model post-training is applied to refine behaviors and unlock new skills across a wide range of recent language models, but open recipes for applying these techniques lag behind proprietary ones. The underlying training data and recipes for post-training are simultaneously the most important pieces of the puzzle and the portion with the least transparency. To bridge this gap, we introduce T\"ULU 3, a family of fully-open state-of-the-art post-trained models, alongside its data, code, and training recipes, serving as a comprehensive guide for modern post-training techniques. T\"ULU 3, which builds on Llama 3.1 base models, achieves results surpassing the instruct versions of Llama 3.1, Qwen 2.5, Mistral, and even closed models such as GPT-4o-mini and Claude 3.5-Haiku. The training algorithms for our models include supervised finetuning (SFT), Direct Preference Optimization (DPO), and a novel method we call Reinforcement Learning with Verifiable Rewards (RLVR). With T\"ULU 3, we introduce a multi-task evaluation scheme for post-training recipes with development and unseen evaluations, standard benchmark implementations, and substantial decontamination of existing open datasets on said benchmarks. We conclude with analysis and discussion of training methods that did not reliably improve performance. In addition to the T\"ULU 3 model weights and demo, we release the complete recipe -- including datasets for diverse core skills, a robust toolkit for data curation and evaluation, the training code and infrastructure, and, most importantly, a detailed report for reproducing and further adapting the T\"ULU 3 approach to more domains.
The Semantic Scholar Open Data Platform
Jan 24, 2023



Abstract:The volume of scientific output is creating an urgent need for automated tools to help scientists keep up with developments in their field. Semantic Scholar (S2) is an open data platform and website aimed at accelerating science by helping scholars discover and understand scientific literature. We combine public and proprietary data sources using state-of-the-art techniques for scholarly PDF content extraction and automatic knowledge graph construction to build the Semantic Scholar Academic Graph, the largest open scientific literature graph to-date, with 200M+ papers, 80M+ authors, 550M+ paper-authorship edges, and 2.4B+ citation edges. The graph includes advanced semantic features such as structurally parsed text, natural language summaries, and vector embeddings. In this paper, we describe the components of the S2 data processing pipeline and the associated APIs offered by the platform. We will update this living document to reflect changes as we add new data offerings and improve existing services.
CORD-19: The Covid-19 Open Research Dataset
Apr 25, 2020
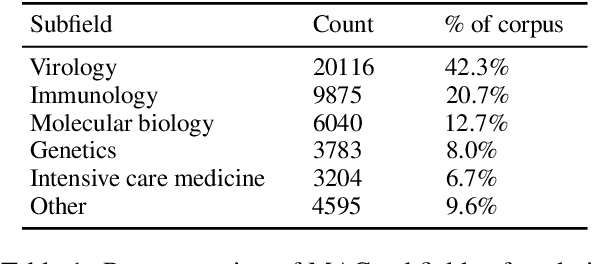


Abstract:The Covid-19 Open Research Dataset (CORD-19) is a growing resource of scientific papers on Covid-19 and related historical coronavirus research. CORD-19 is designed to facilitate the development of text mining and information retrieval systems over its rich collection of metadata and structured full text papers. Since its release, CORD-19 has been downloaded over 75K times and has served as the basis of many Covid-19 text mining and discovery systems. In this article, we describe the mechanics of dataset construction, highlighting challenges and key design decisions, provide an overview of how CORD-19 has been used, and preview tools and upcoming shared tasks built around the dataset. We hope this resource will continue to bring together the computing community, biomedical experts, and policy makers in the search for effective treatments and management policies for Covid-19.
Ontology Alignment in the Biomedical Domain Using Entity Definitions and Context
Jun 20, 2018



Abstract:Ontology alignment is the task of identifying semantically equivalent entities from two given ontologies. Different ontologies have different representations of the same entity, resulting in a need to de-duplicate entities when merging ontologies. We propose a method for enriching entities in an ontology with external definition and context information, and use this additional information for ontology alignment. We develop a neural architecture capable of encoding the additional information when available, and show that the addition of external data results in an F1-score of 0.69 on the Ontology Alignment Evaluation Initiative (OAEI) largebio SNOMED-NCI subtask, comparable with the entity-level matchers in a SOTA system.
Construction of the Literature Graph in Semantic Scholar
May 06, 2018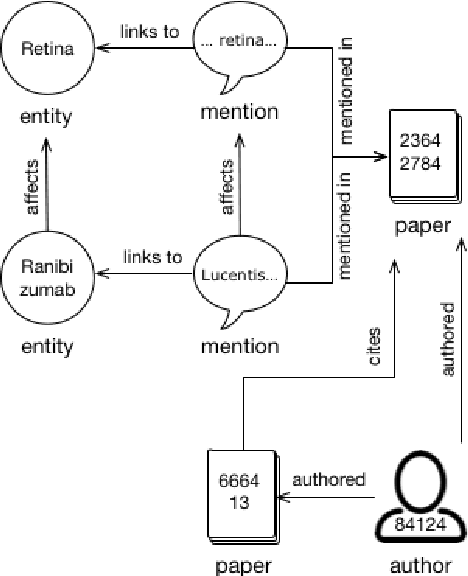
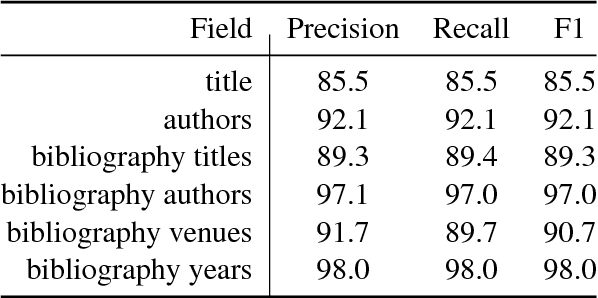
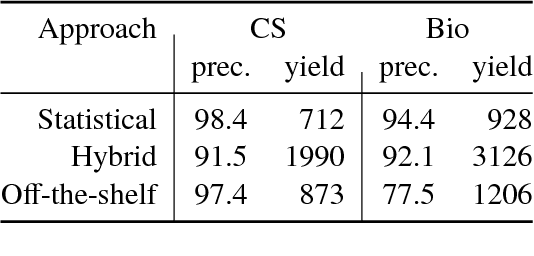
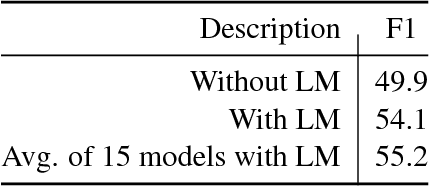
Abstract:We describe a deployed scalable system for organizing published scientific literature into a heterogeneous graph to facilitate algorithmic manipulation and discovery. The resulting literature graph consists of more than 280M nodes, representing papers, authors, entities and various interactions between them (e.g., authorships, citations, entity mentions). We reduce literature graph construction into familiar NLP tasks (e.g., entity extraction and linking), point out research challenges due to differences from standard formulations of these tasks, and report empirical results for each task. The methods described in this paper are used to enable semantic features in www.semanticscholar.org
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge