Jiangjiang Yang
TÜLU 3: Pushing Frontiers in Open Language Model Post-Training
Nov 22, 2024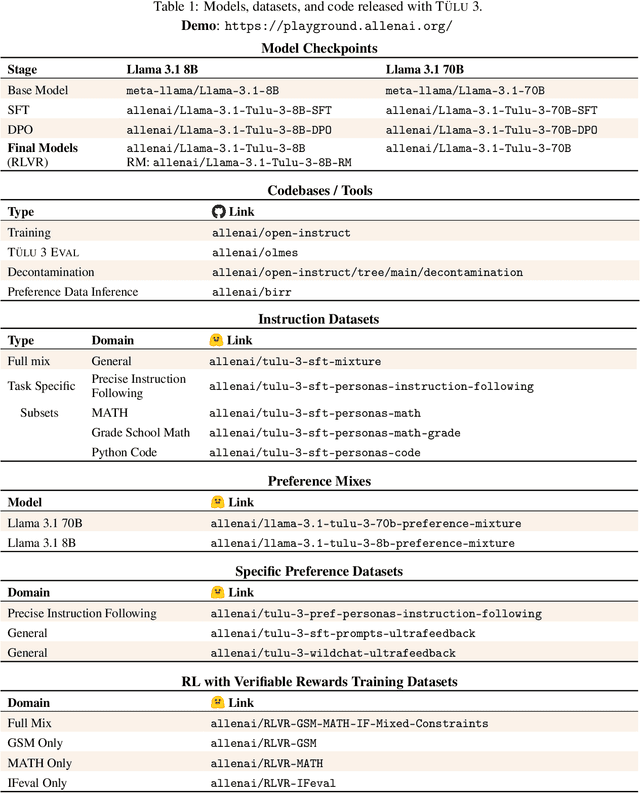



Abstract:Language model post-training is applied to refine behaviors and unlock new skills across a wide range of recent language models, but open recipes for applying these techniques lag behind proprietary ones. The underlying training data and recipes for post-training are simultaneously the most important pieces of the puzzle and the portion with the least transparency. To bridge this gap, we introduce T\"ULU 3, a family of fully-open state-of-the-art post-trained models, alongside its data, code, and training recipes, serving as a comprehensive guide for modern post-training techniques. T\"ULU 3, which builds on Llama 3.1 base models, achieves results surpassing the instruct versions of Llama 3.1, Qwen 2.5, Mistral, and even closed models such as GPT-4o-mini and Claude 3.5-Haiku. The training algorithms for our models include supervised finetuning (SFT), Direct Preference Optimization (DPO), and a novel method we call Reinforcement Learning with Verifiable Rewards (RLVR). With T\"ULU 3, we introduce a multi-task evaluation scheme for post-training recipes with development and unseen evaluations, standard benchmark implementations, and substantial decontamination of existing open datasets on said benchmarks. We conclude with analysis and discussion of training methods that did not reliably improve performance. In addition to the T\"ULU 3 model weights and demo, we release the complete recipe -- including datasets for diverse core skills, a robust toolkit for data curation and evaluation, the training code and infrastructure, and, most importantly, a detailed report for reproducing and further adapting the T\"ULU 3 approach to more domains.
The Semantic Reader Project: Augmenting Scholarly Documents through AI-Powered Interactive Reading Interfaces
Mar 25, 2023
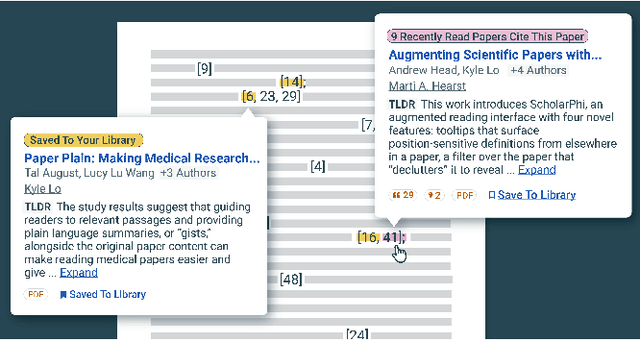
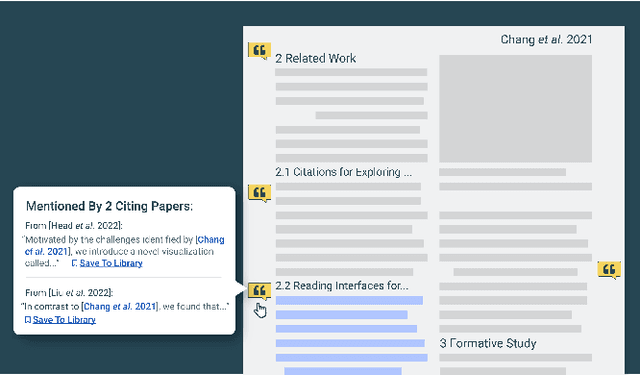
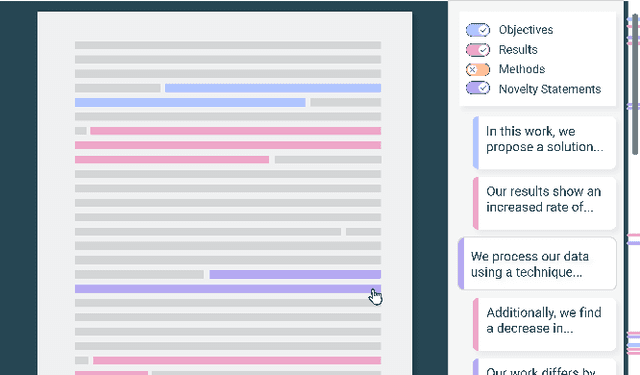
Abstract:Scholarly publications are key to the transfer of knowledge from scholars to others. However, research papers are information-dense, and as the volume of the scientific literature grows, the need for new technology to support the reading process grows. In contrast to the process of finding papers, which has been transformed by Internet technology, the experience of reading research papers has changed little in decades. The PDF format for sharing research papers is widely used due to its portability, but it has significant downsides including: static content, poor accessibility for low-vision readers, and difficulty reading on mobile devices. This paper explores the question "Can recent advances in AI and HCI power intelligent, interactive, and accessible reading interfaces -- even for legacy PDFs?" We describe the Semantic Reader Project, a collaborative effort across multiple institutions to explore automatic creation of dynamic reading interfaces for research papers. Through this project, we've developed ten research prototype interfaces and conducted usability studies with more than 300 participants and real-world users showing improved reading experiences for scholars. We've also released a production reading interface for research papers that will incorporate the best features as they mature. We structure this paper around challenges scholars and the public face when reading research papers -- Discovery, Efficiency, Comprehension, Synthesis, and Accessibility -- and present an overview of our progress and remaining open challenges.
The Semantic Scholar Open Data Platform
Jan 24, 2023



Abstract:The volume of scientific output is creating an urgent need for automated tools to help scientists keep up with developments in their field. Semantic Scholar (S2) is an open data platform and website aimed at accelerating science by helping scholars discover and understand scientific literature. We combine public and proprietary data sources using state-of-the-art techniques for scholarly PDF content extraction and automatic knowledge graph construction to build the Semantic Scholar Academic Graph, the largest open scientific literature graph to-date, with 200M+ papers, 80M+ authors, 550M+ paper-authorship edges, and 2.4B+ citation edges. The graph includes advanced semantic features such as structurally parsed text, natural language summaries, and vector embeddings. In this paper, we describe the components of the S2 data processing pipeline and the associated APIs offered by the platform. We will update this living document to reflect changes as we add new data offerings and improve existing services.
From Who You Know to What You Read: Augmenting Scientific Recommendations with Implicit Social Networks
Apr 21, 2022
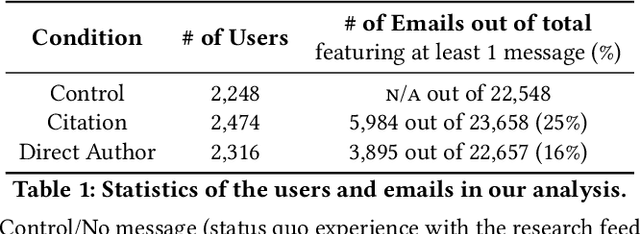

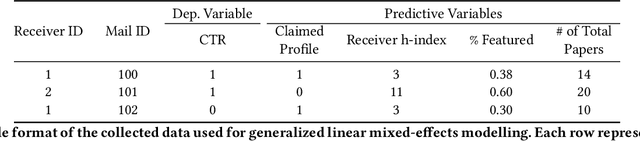
Abstract:The ever-increasing pace of scientific publication necessitates methods for quickly identifying relevant papers. While neural recommenders trained on user interests can help, they still result in long, monotonous lists of suggested papers. To improve the discovery experience we introduce multiple new methods for \em augmenting recommendations with textual relevance messages that highlight knowledge-graph connections between recommended papers and a user's publication and interaction history. We explore associations mediated by author entities and those using citations alone. In a large-scale, real-world study, we show how our approach significantly increases engagement -- and future engagement when mediated by authors -- without introducing bias towards highly-cited authors. To expand message coverage for users with less publication or interaction history, we develop a novel method that highlights connections with proxy authors of interest to users and evaluate it in a controlled lab study. Finally, we synthesize design implications for future graph-based messages.
CORD-19: The Covid-19 Open Research Dataset
Apr 25, 2020
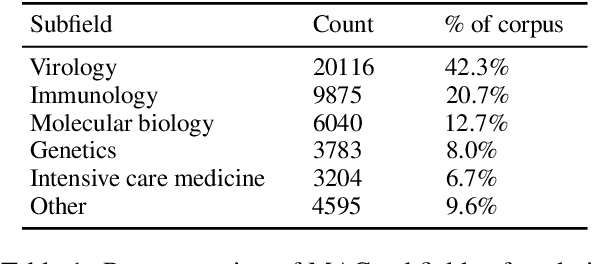


Abstract:The Covid-19 Open Research Dataset (CORD-19) is a growing resource of scientific papers on Covid-19 and related historical coronavirus research. CORD-19 is designed to facilitate the development of text mining and information retrieval systems over its rich collection of metadata and structured full text papers. Since its release, CORD-19 has been downloaded over 75K times and has served as the basis of many Covid-19 text mining and discovery systems. In this article, we describe the mechanics of dataset construction, highlighting challenges and key design decisions, provide an overview of how CORD-19 has been used, and preview tools and upcoming shared tasks built around the dataset. We hope this resource will continue to bring together the computing community, biomedical experts, and policy makers in the search for effective treatments and management policies for Covid-19.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge