Sophie Johnson
ACCoRD: A Multi-Document Approach to Generating Diverse Descriptions of Scientific Concepts
May 14, 2022
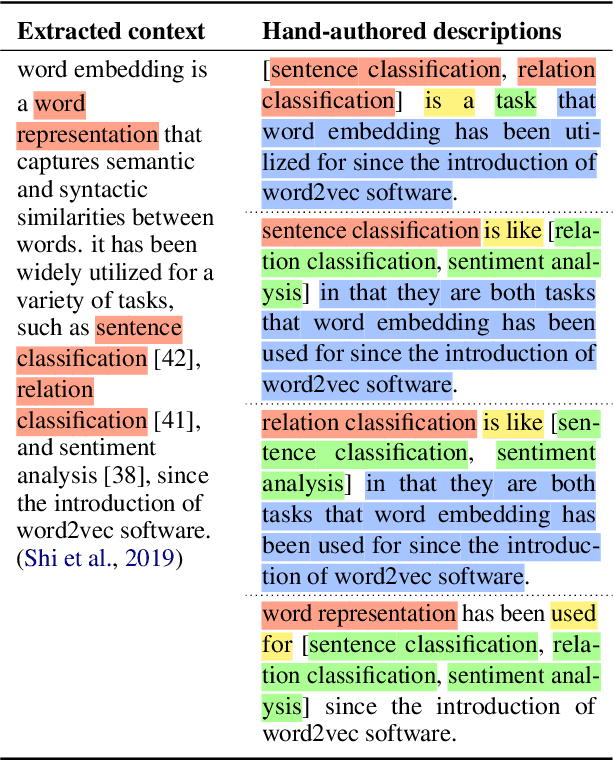

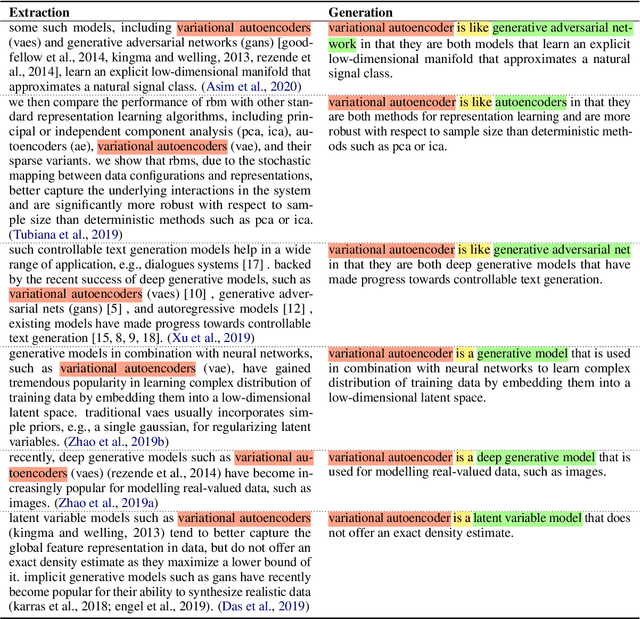
Abstract:Systems that can automatically define unfamiliar terms hold the promise of improving the accessibility of scientific texts, especially for readers who may lack prerequisite background knowledge. However, current systems assume a single "best" description per concept, which fails to account for the many potentially useful ways a concept can be described. We present ACCoRD, an end-to-end system tackling the novel task of generating sets of descriptions of scientific concepts. Our system takes advantage of the myriad ways a concept is mentioned across the scientific literature to produce distinct, diverse descriptions of target scientific concepts in terms of different reference concepts. To support research on the task, we release an expert-annotated resource, the ACCoRD corpus, which includes 1,275 labeled contexts and 1,787 hand-authored concept descriptions. We conduct a user study demonstrating that (1) users prefer descriptions produced by our end-to-end system, and (2) users prefer multiple descriptions to a single "best" description.
A Search Engine for Discovery of Scientific Challenges and Directions
Sep 10, 2021



Abstract:Keeping track of scientific challenges, advances and emerging directions is a fundamental part of research. However, researchers face a flood of papers that hinders discovery of important knowledge. In biomedicine, this directly impacts human lives. To address this problem, we present a novel task of extraction and search of scientific challenges and directions, to facilitate rapid knowledge discovery. We construct and release an expert-annotated corpus of texts sampled from full-length papers, labeled with novel semantic categories that generalize across many types of challenges and directions. We focus on a large corpus of interdisciplinary work relating to the COVID-19 pandemic, ranging from biomedicine to areas such as AI and economics. We apply a model trained on our data to identify challenges and directions across the corpus and build a dedicated search engine. In experiments with 19 researchers and clinicians using our system, we outperform a popular scientific search engine in assisting knowledge discovery. Finally, we show that models trained on our resource generalize to the wider biomedical domain and to AI papers, highlighting its broad utility. We make our data, model and search engine publicly available. https://challenges.apps.allenai.org/
MultiCite: Modeling realistic citations requires moving beyond the single-sentence single-label setting
Aug 01, 2021



Abstract:Citation context analysis (CCA) is an important task in natural language processing that studies how and why scholars discuss each others' work. Despite decades of study, traditional frameworks for CCA have largely relied on overly-simplistic assumptions of how authors cite, which ignore several important phenomena. For instance, scholarly papers often contain rich discussions of cited work that span multiple sentences and express multiple intents concurrently. Yet, CCA is typically approached as a single-sentence, single-label classification task, and thus existing datasets fail to capture this interesting discourse. In our work, we address this research gap by proposing a novel framework for CCA as a document-level context extraction and labeling task. We release MultiCite, a new dataset of 12,653 citation contexts from over 1,200 computational linguistics papers. Not only is it the largest collection of expert-annotated citation contexts to-date, MultiCite contains multi-sentence, multi-label citation contexts within full paper texts. Finally, we demonstrate how our dataset, while still usable for training classic CCA models, also supports the development of new types of models for CCA beyond fixed-width text classification. We release our code and dataset at https://github.com/allenai/multicite.
SciCo: Hierarchical Cross-Document Coreference for Scientific Concepts
Apr 18, 2021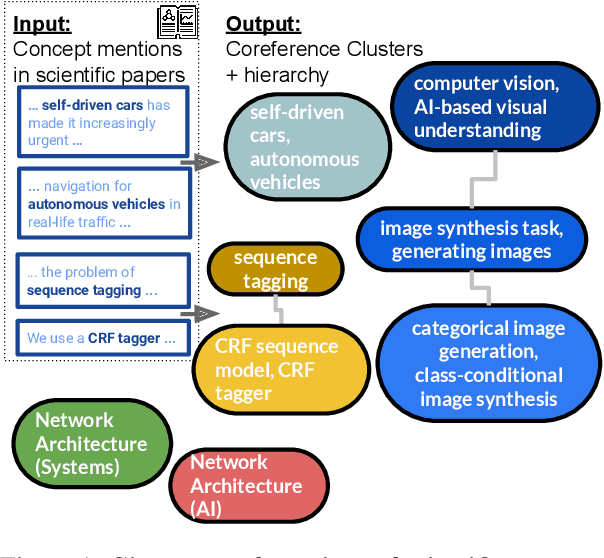

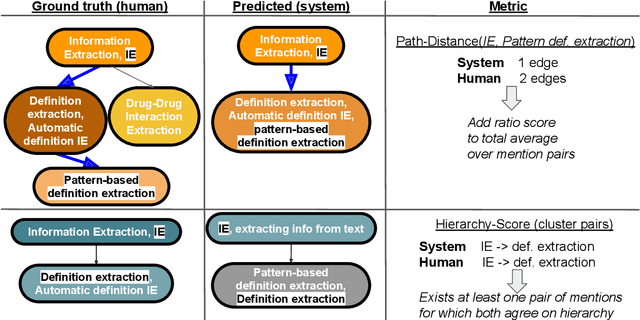
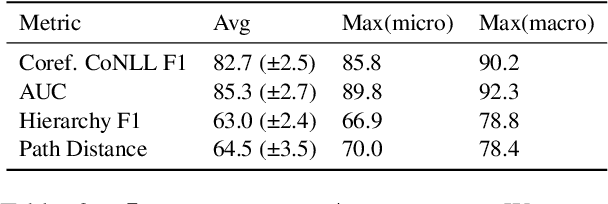
Abstract:Determining coreference of concept mentions across multiple documents is fundamental for natural language understanding. Work on cross-document coreference resolution (CDCR) typically considers mentions of events in the news, which do not often involve abstract technical concepts that are prevalent in science and technology. These complex concepts take diverse or ambiguous forms and have many hierarchical levels of granularity (e.g., tasks and subtasks), posing challenges for CDCR. We present a new task of hierarchical CDCR for concepts in scientific papers, with the goal of jointly inferring coreference clusters and hierarchy between them. We create SciCo, an expert-annotated dataset for this task, which is 3X larger than the prominent ECB+ resource. We find that tackling both coreference and hierarchy at once outperforms disjoint models, which we hope will spur development of joint models for SciCo.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge