Yusong Wang
BMAM: Brain-inspired Multi-Agent Memory Framework
Jan 28, 2026Abstract:Language-model-based agents operating over extended interaction horizons face persistent challenges in preserving temporally grounded information and maintaining behavioral consistency across sessions, a failure mode we term soul erosion. We present BMAM (Brain-inspired Multi-Agent Memory), a general-purpose memory architecture that models agent memory as a set of functionally specialized subsystems rather than a single unstructured store. Inspired by cognitive memory systems, BMAM decomposes memory into episodic, semantic, salience-aware, and control-oriented components that operate at complementary time scales. To support long-horizon reasoning, BMAM organizes episodic memories along explicit timelines and retrieves evidence by fusing multiple complementary signals. Experiments on the LoCoMo benchmark show that BMAM achieves 78.45 percent accuracy under the standard long-horizon evaluation setting, and ablation analyses confirm that the hippocampus-inspired episodic memory subsystem plays a critical role in temporal reasoning.
MMPG: MoE-based Adaptive Multi-Perspective Graph Fusion for Protein Representation Learning
Jan 15, 2026Abstract:Graph Neural Networks (GNNs) have been widely adopted for Protein Representation Learning (PRL), as residue interaction networks can be naturally represented as graphs. Current GNN-based PRL methods typically rely on single-perspective graph construction strategies, which capture partial properties of residue interactions, resulting in incomplete protein representations. To address this limitation, we propose MMPG, a framework that constructs protein graphs from multiple perspectives and adaptively fuses them via Mixture of Experts (MoE) for PRL. MMPG constructs graphs from physical, chemical, and geometric perspectives to characterize different properties of residue interactions. To capture both perspective-specific features and their synergies, we develop an MoE module, which dynamically routes perspectives to specialized experts, where experts learn intrinsic features and cross-perspective interactions. We quantitatively verify that MoE automatically specializes experts in modeling distinct levels of interaction from individual representations, to pairwise inter-perspective synergies, and ultimately to a global consensus across all perspectives. Through integrating this multi-level information, MMPG produces superior protein representations and achieves advanced performance on four different downstream protein tasks.
MCGM: Multi-stage Clustered Global Modeling for Long-range Interactions in Molecules
Sep 26, 2025Abstract:Geometric graph neural networks (GNNs) excel at capturing molecular geometry, yet their locality-biased message passing hampers the modeling of long-range interactions. Current solutions have fundamental limitations: extending cutoff radii causes computational costs to scale cubically with distance; physics-inspired kernels (e.g., Coulomb, dispersion) are often system-specific and lack generality; Fourier-space methods require careful tuning of multiple parameters (e.g., mesh size, k-space cutoff) with added computational overhead. We introduce Multi-stage Clustered Global Modeling (MCGM), a lightweight, plug-and-play module that endows geometric GNNs with hierarchical global context through efficient clustering operations. MCGM builds a multi-resolution hierarchy of atomic clusters, distills global information via dynamic hierarchical clustering, and propagates this context back through learned transformations, ultimately reinforcing atomic features via residual connections. Seamlessly integrated into four diverse backbone architectures, MCGM reduces OE62 energy prediction error by an average of 26.2%. On AQM, MCGM achieves state-of-the-art accuracy (17.0 meV for energy, 4.9 meV/{\AA} for forces) while using 20% fewer parameters than Neural P3M. Code will be made available upon acceptance.
GenColor: Generative Color-Concept Association in Visual Design
Mar 05, 2025Abstract:Existing approaches for color-concept association typically rely on query-based image referencing, and color extraction from image references. However, these approaches are effective only for common concepts, and are vulnerable to unstable image referencing and varying image conditions. Our formative study with designers underscores the need for primary-accent color compositions and context-dependent colors (e.g., 'clear' vs. 'polluted' sky) in design. In response, we introduce a generative approach for mining semantically resonant colors leveraging images generated by text-to-image models. Our insight is that contemporary text-to-image models can resemble visual patterns from large-scale real-world data. The framework comprises three stages: concept instancing produces generative samples using diffusion models, text-guided image segmentation identifies concept-relevant regions within the image, and color association extracts primarily accompanied by accent colors. Quantitative comparisons with expert designs validate our approach's effectiveness, and we demonstrate the applicability through cases in various design scenarios and a gallery.
Tokenizing 3D Molecule Structure with Quantized Spherical Coordinates
Dec 02, 2024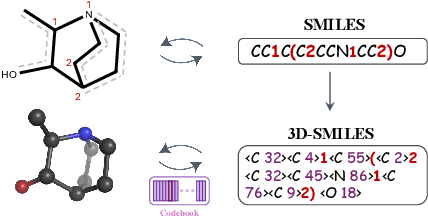
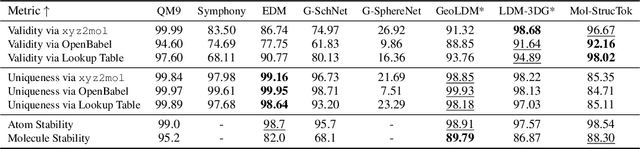
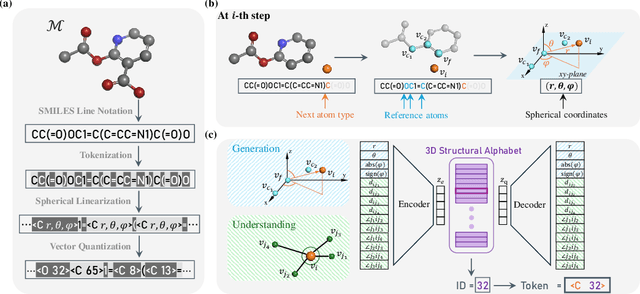
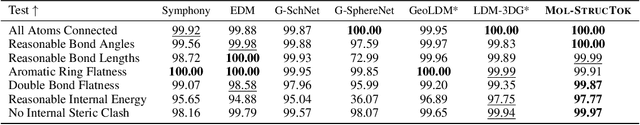
Abstract:The application of language models (LMs) to molecular structure generation using line notations such as SMILES and SELFIES has been well-established in the field of cheminformatics. However, extending these models to generate 3D molecular structures presents significant challenges. Two primary obstacles emerge: (1) the difficulty in designing a 3D line notation that ensures SE(3)-invariant atomic coordinates, and (2) the non-trivial task of tokenizing continuous coordinates for use in LMs, which inherently require discrete inputs. To address these challenges, we propose Mol-StrucTok, a novel method for tokenizing 3D molecular structures. Our approach comprises two key innovations: (1) We design a line notation for 3D molecules by extracting local atomic coordinates in a spherical coordinate system. This notation builds upon existing 2D line notations and remains agnostic to their specific forms, ensuring compatibility with various molecular representation schemes. (2) We employ a Vector Quantized Variational Autoencoder (VQ-VAE) to tokenize these coordinates, treating them as generation descriptors. To further enhance the representation, we incorporate neighborhood bond lengths and bond angles as understanding descriptors. Leveraging this tokenization framework, we train a GPT-2 style model for 3D molecular generation tasks. Results demonstrate strong performance with significantly faster generation speeds and competitive chemical stability compared to previous methods. Further, by integrating our learned discrete representations into Graphormer model for property prediction on QM9 dataset, Mol-StrucTok reveals consistent improvements across various molecular properties, underscoring the versatility and robustness of our approach.
Brain-inspired continual pre-trained learner via silent synaptic consolidation
Oct 08, 2024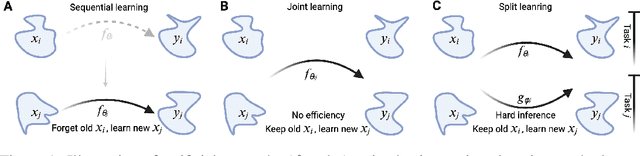
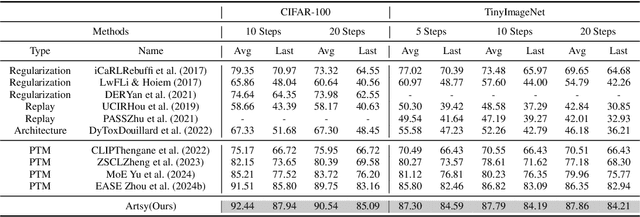
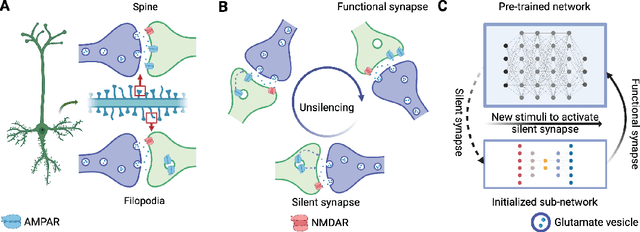
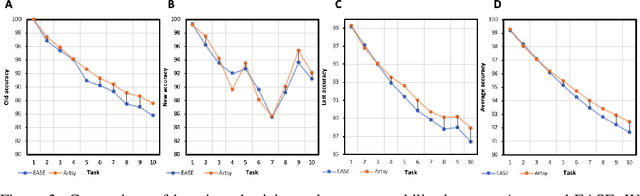
Abstract:Pre-trained models have demonstrated impressive generalization capabilities, yet they remain vulnerable to catastrophic forgetting when incrementally trained on new tasks. Existing architecture-based strategies encounter two primary challenges: 1) Integrating a pre-trained network with a trainable sub-network complicates the delicate balance between learning plasticity and memory stability across evolving tasks during learning. 2) The absence of robust interconnections between pre-trained networks and various sub-networks limits the effective retrieval of pertinent information during inference. In this study, we introduce the Artsy, inspired by the activation mechanisms of silent synapses via spike-timing-dependent plasticity observed in mature brains, to enhance the continual learning capabilities of pre-trained models. The Artsy integrates two key components: During training, the Artsy mimics mature brain dynamics by maintaining memory stability for previously learned knowledge within the pre-trained network while simultaneously promoting learning plasticity in task-specific sub-networks. During inference, artificial silent and functional synapses are utilized to establish precise connections between the pre-synaptic neurons in the pre-trained network and the post-synaptic neurons in the sub-networks, facilitated through synaptic consolidation, thereby enabling effective extraction of relevant information from test samples. Comprehensive experimental evaluations reveal that our model significantly outperforms conventional methods on class-incremental learning tasks, while also providing enhanced biological interpretability for architecture-based approaches. Moreover, we propose that the Artsy offers a promising avenue for simulating biological synaptic mechanisms, potentially advancing our understanding of neural plasticity in both artificial and biological systems.
Neural P$^3$M: A Long-Range Interaction Modeling Enhancer for Geometric GNNs
Sep 26, 2024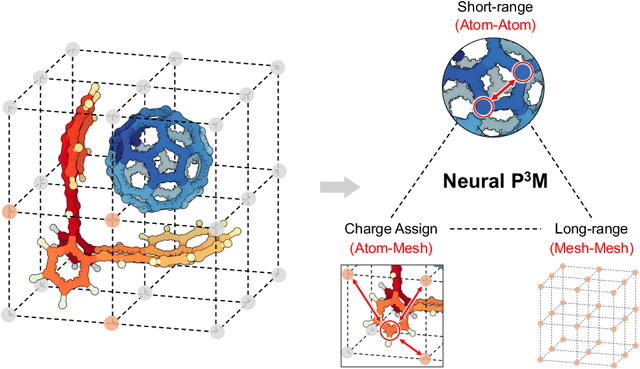
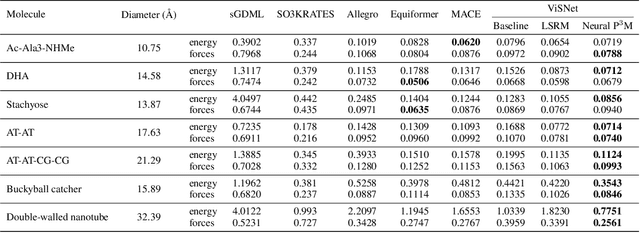
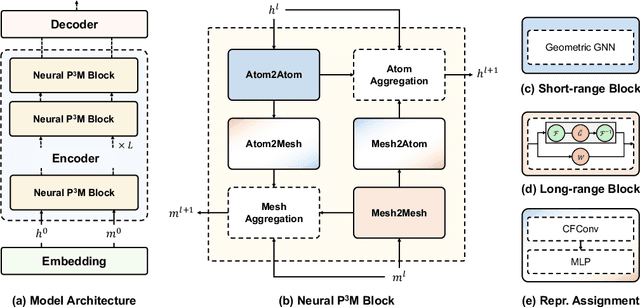
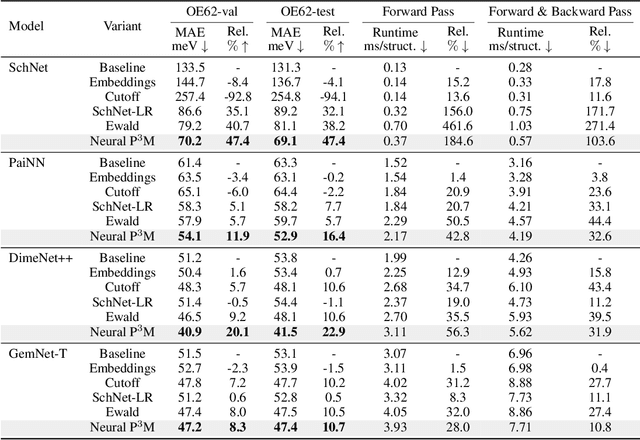
Abstract:Geometric graph neural networks (GNNs) have emerged as powerful tools for modeling molecular geometry. However, they encounter limitations in effectively capturing long-range interactions in large molecular systems. To address this challenge, we introduce Neural P$^3$M, a versatile enhancer of geometric GNNs to expand the scope of their capabilities by incorporating mesh points alongside atoms and reimaging traditional mathematical operations in a trainable manner. Neural P$^3$M exhibits flexibility across a wide range of molecular systems and demonstrates remarkable accuracy in predicting energies and forces, outperforming on benchmarks such as the MD22 dataset. It also achieves an average improvement of 22% on the OE62 dataset while integrating with various architectures.
Advancing Cross-domain Discriminability in Continual Learning of Vison-Language Models
Jun 27, 2024



Abstract:Continual learning (CL) with Vision-Language Models (VLMs) has overcome the constraints of traditional CL, which only focuses on previously encountered classes. During the CL of VLMs, we need not only to prevent the catastrophic forgetting on incrementally learned knowledge but also to preserve the zero-shot ability of VLMs. However, existing methods require additional reference datasets to maintain such zero-shot ability and rely on domain-identity hints to classify images across different domains. In this study, we propose Regression-based Analytic Incremental Learning (RAIL), which utilizes a recursive ridge regression-based adapter to learn from a sequence of domains in a non-forgetting manner and decouple the cross-domain correlations by projecting features to a higher-dimensional space. Cooperating with a training-free fusion module, RAIL absolutely preserves the VLM's zero-shot ability on unseen domains without any reference data. Additionally, we introduce Cross-domain Task-Agnostic Incremental Learning (X-TAIL) setting. In this setting, a CL learner is required to incrementally learn from multiple domains and classify test images from both seen and unseen domains without any domain-identity hint. We theoretically prove RAIL's absolute memorization on incrementally learned domains. Experiment results affirm RAIL's state-of-the-art performance in both X-TAIL and existing Multi-domain Task-Incremental Learning settings. The code will be released upon acceptance.
VBIM-Net: Variational Born Iterative Network for Inverse Scattering Problems
May 29, 2024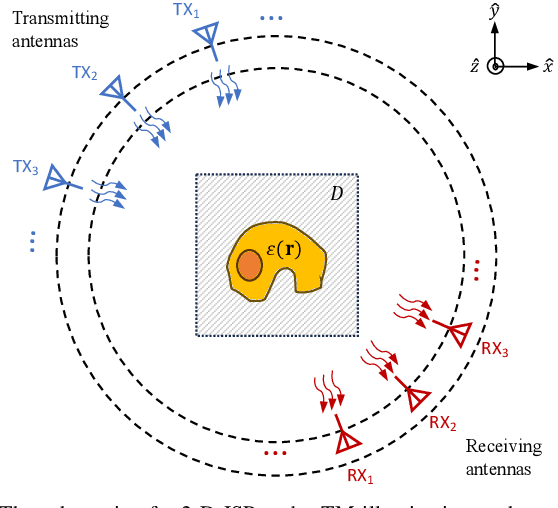

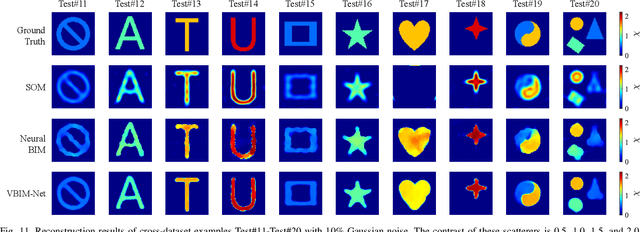

Abstract:Recently, studies have shown the potential of integrating field-type iterative methods with deep learning (DL) techniques in solving inverse scattering problems (ISPs). In this article, we propose a novel Variational Born Iterative Network, namely, VBIM-Net, to solve the full-wave ISPs with significantly improved flexibility and inversion quality. The proposed VBIM-Net emulates the alternating updates of the total electric field and the contrast in the variational Born iterative method (VBIM) by multiple layers of subnetworks. We embed the calculation of the contrast variation into each of the subnetworks, converting the scattered field residual into an approximate contrast variation and then enhancing it by a U-Net, thus avoiding the requirement of matched measurement dimension and grid resolution as in existing approaches. The total field and contrast of each layer's output is supervised in the loss function of VBIM-Net, which guarantees the physical interpretability of variables of the subnetworks. In addition, we design a training scheme with extra noise to enhance the model's stability. Extensive numerical results on synthetic and experimental data both verify the inversion quality, generalization ability, and robustness of the proposed VBIM-Net. This work may provide some new inspiration for the design of efficient field-type DL schemes.
F$^3$low: Frame-to-Frame Coarse-grained Molecular Dynamics with SE Guided Flow Matching
May 01, 2024



Abstract:Molecular dynamics (MD) is a crucial technique for simulating biological systems, enabling the exploration of their dynamic nature and fostering an understanding of their functions and properties. To address exploration inefficiency, emerging enhanced sampling approaches like coarse-graining (CG) and generative models have been employed. In this work, we propose a \underline{Frame-to-Frame} generative model with guided \underline{Flow}-matching (F$3$low) for enhanced sampling, which (a) extends the domain of CG modeling to the SE(3) Riemannian manifold; (b) retreating CGMD simulations as autoregressively sampling guided by the former frame via flow-matching models; (c) targets the protein backbone, offering improved insights into secondary structure formation and intricate folding pathways. Compared to previous methods, F$3$low allows for broader exploration of conformational space. The ability to rapidly generate diverse conformations via force-free generative paradigm on SE(3) paves the way toward efficient enhanced sampling methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge