Yuan Qi
Fudan University
Uncovering Modality Discrepancy and Generalization Illusion for General-Purpose 3D Medical Segmentation
Feb 07, 2026Abstract:While emerging 3D medical foundation models are envisioned as versatile tools with offer general-purpose capabilities, their validation remains largely confined to regional and structural imaging, leaving a significant modality discrepancy unexplored. To provide a rigorous and objective assessment, we curate the UMD dataset comprising 490 whole-body PET/CT and 464 whole-body PET/MRI scans ($\sim$675k 2D images, $\sim$12k 3D organ annotations) and conduct a thorough and comprehensive evaluation of representative 3D segmentation foundation models. Through intra-subject controlled comparisons of paired scans, we isolate imaging modality as the primary independent variable to evaluate model robustness in real-world applications. Our evaluation reveals a stark discrepancy between literature-reported benchmarks and real-world efficacy, particularly when transitioning from structural to functional domains. Such systemic failures underscore that current 3D foundation models are far from achieving truly general-purpose status, necessitating a paradigm shift toward multi-modal training and evaluation to bridge the gap between idealized benchmarking and comprehensive clinical utility. This dataset and analysis establish a foundational cornerstone for future research to develop truly modality-agnostic medical foundation models.
Structure-based RNA Design by Step-wise Optimization of Latent Diffusion Model
Jan 27, 2026Abstract:RNA inverse folding, designing sequences to form specific 3D structures, is critical for therapeutics, gene regulation, and synthetic biology. Current methods, focused on sequence recovery, struggle to address structural objectives like secondary structure consistency (SS), minimum free energy (MFE), and local distance difference test (LDDT), leading to suboptimal structural accuracy. To tackle this, we propose a reinforcement learning (RL) framework integrated with a latent diffusion model (LDM). Drawing inspiration from the success of diffusion models in RNA inverse folding, which adeptly model complex sequence-structure interactions, we develop an LDM incorporating pre-trained RNA-FM embeddings from a large-scale RNA model. These embeddings capture co-evolutionary patterns, markedly improving sequence recovery accuracy. However, existing approaches, including diffusion-based methods, cannot effectively handle non-differentiable structural objectives. By contrast, RL excels in this task by using policy-driven reward optimization to navigate complex, non-gradient-based objectives, offering a significant advantage over traditional methods. In summary, we propose the Step-wise Optimization of Latent Diffusion Model (SOLD), a novel RL framework that optimizes single-step noise without sampling the full diffusion trajectory, achieving efficient refinement of multiple structural objectives. Experimental results demonstrate SOLD surpasses its LDM baseline and state-of-the-art methods across all metrics, establishing a robust framework for RNA inverse folding with profound implications for biotechnological and therapeutic applications.
A Pre-trained Reaction Embedding Descriptor Capturing Bond Transformation Patterns
Jan 07, 2026Abstract:With the rise of data-driven reaction prediction models, effective reaction descriptors are crucial for bridging the gap between real-world chemistry and digital representations. However, general-purpose, reaction-wise descriptors remain scarce. This study introduces RXNEmb, a novel reaction-level descriptor derived from RXNGraphormer, a model pre-trained to distinguish real reactions from fictitious ones with erroneous bond changes, thereby learning intrinsic bond formation and cleavage patterns. We demonstrate its utility by data-driven re-clustering of the USPTO-50k dataset, yielding a classification that more directly reflects bond-change similarities than rule-based categories. Combined with dimensionality reduction, RXNEmb enables visualization of reaction space diversity. Furthermore, attention weight analysis reveals the model's focus on chemically critical sites, providing mechanistic insight. RXNEmb serves as a powerful, interpretable tool for reaction fingerprinting and analysis, paving the way for more data-centric approaches in reaction analysis and discovery.
CTkvr: KV Cache Retrieval for Long-Context LLMs via Centroid then Token Indexing
Dec 17, 2025Abstract:Large language models (LLMs) are increasingly applied in long-context scenarios such as multi-turn conversations. However, long contexts pose significant challenges for inference efficiency, including high memory overhead from Key-Value (KV) cache and increased latency due to excessive memory accesses. Recent methods for dynamic KV selection struggle with trade-offs: block-level indexing degrades accuracy by retrieving irrelevant KV entries, while token-level indexing incurs high latency from inefficient retrieval mechanisms. In this paper, we propose CTKVR, a novel centroid-then-token KV retrieval scheme that addresses these limitations. CTKVR leverages a key observation: query vectors adjacent in position exhibit high similarity after Rotary Position Embedding (RoPE) and share most of their top-k KV cache entries. Based on this insight, CTKVR employs a two-stage retrieval strategy: lightweight centroids are precomputed during prefilling for centroid-grained indexing, followed by token-level refinement for precise KV retrieval. This approach balances retrieval efficiency and accuracy. To further enhance performance, we implement an optimized system for indexing construction and search using CPU-GPU co-execution. Experimentally, CTKVR achieves superior performance across multiple benchmarks with less than 1% accuracy degradation. Meanwhile, CTKVR delivers 3 times and 4 times throughput speedups on Llama-3-8B and Yi-9B at 96K context length across diverse GPU hardware.
Finding Kissing Numbers with Game-theoretic Reinforcement Learning
Nov 17, 2025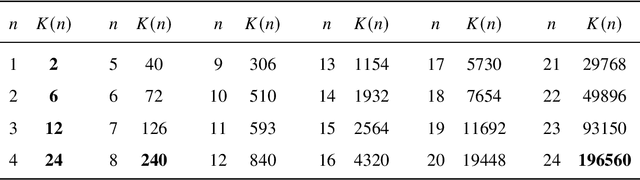
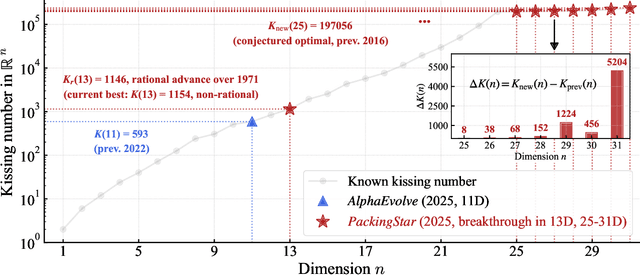
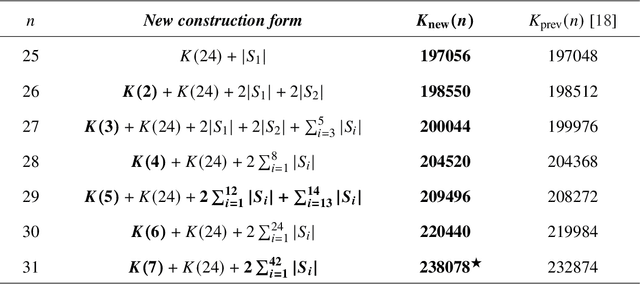
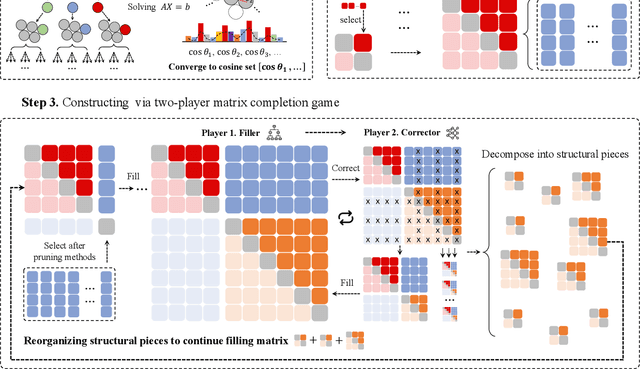
Abstract:Since Isaac Newton first studied the Kissing Number Problem in 1694, determining the maximal number of non-overlapping spheres around a central sphere has remained a fundamental challenge. This problem represents the local analogue of Hilbert's 18th problem on sphere packing, bridging geometry, number theory, and information theory. Although significant progress has been made through lattices and codes, the irregularities of high-dimensional geometry and exponentially growing combinatorial complexity beyond 8 dimensions, which exceeds the complexity of Go game, limit the scalability of existing methods. Here we model this problem as a two-player matrix completion game and train the game-theoretic reinforcement learning system, PackingStar, to efficiently explore high-dimensional spaces. The matrix entries represent pairwise cosines of sphere center vectors; one player fills entries while another corrects suboptimal ones, jointly maximizing the matrix size, corresponding to the kissing number. This cooperative dynamics substantially improves sample quality, making the extremely large spaces tractable. PackingStar reproduces previous configurations and surpasses all human-known records from dimensions 25 to 31, with the configuration in 25 dimensions geometrically corresponding to the Leech lattice and suggesting possible optimality. It achieves the first breakthrough beyond rational structures from 1971 in 13 dimensions and discovers over 6000 new structures in 14 and other dimensions. These results demonstrate AI's power to explore high-dimensional spaces beyond human intuition and open new pathways for the Kissing Number Problem and broader geometry problems.
The Choice of Divergence: A Neglected Key to Mitigating Diversity Collapse in Reinforcement Learning with Verifiable Reward
Sep 09, 2025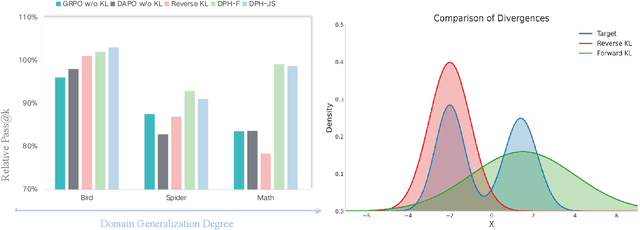
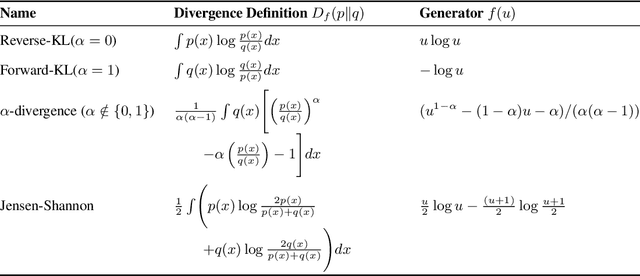
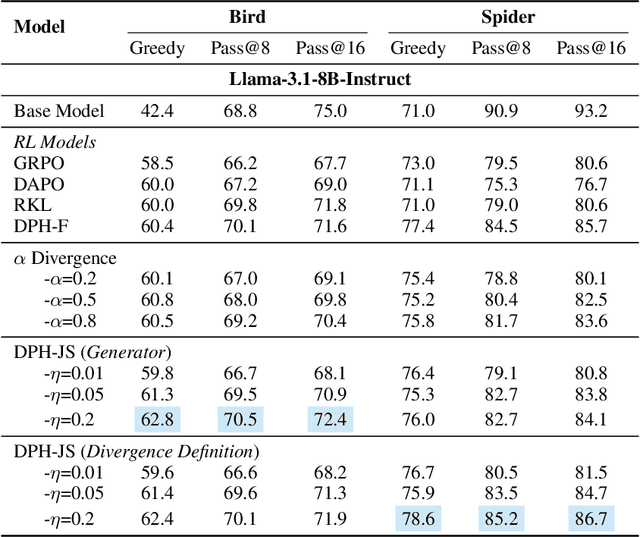
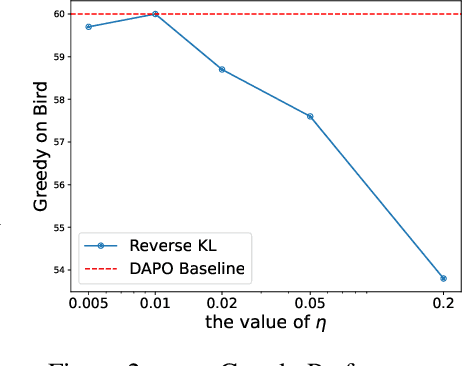
Abstract:A central paradox in fine-tuning Large Language Models (LLMs) with Reinforcement Learning with Verifiable Reward (RLVR) is the frequent degradation of multi-attempt performance (Pass@k) despite improvements in single-attempt accuracy (Pass@1). This is often accompanied by catastrophic forgetting, where models lose previously acquired skills. While various methods have been proposed, the choice and function of the divergence term have been surprisingly unexamined as a proactive solution. We argue that standard RLVR objectives -- both those using the mode-seeking reverse KL-divergence and those forgoing a divergence term entirely -- lack a crucial mechanism for knowledge retention. The reverse-KL actively accelerates this decay by narrowing the policy, while its absence provides no safeguard against the model drifting from its diverse knowledge base. We propose a fundamental shift in perspective: using the divergence term itself as the solution. Our framework, Diversity-Preserving Hybrid RL (DPH-RL), leverages mass-covering f-divergences (like forward-KL and JS-divergence) to function as a rehearsal mechanism. By continuously referencing the initial policy, this approach forces the model to maintain broad solution coverage. Extensive experiments on math and SQL generation demonstrate that DPH-RL not only resolves the Pass@k degradation but improves both Pass@1 and Pass@k in- and out-of-domain. Additionally, DPH-RL is more training-efficient because it computes f-divergence using generator functions, requiring only sampling from the initial policy and no online reference model. Our work highlights a crucial, overlooked axis for improving RLVR, demonstrating that the proper selection of a divergence measure is a powerful tool for building more general and diverse reasoning models.
PET2Rep: Towards Vision-Language Model-Drived Automated Radiology Report Generation for Positron Emission Tomography
Aug 06, 2025Abstract:Positron emission tomography (PET) is a cornerstone of modern oncologic and neurologic imaging, distinguished by its unique ability to illuminate dynamic metabolic processes that transcend the anatomical focus of traditional imaging technologies. Radiology reports are essential for clinical decision making, yet their manual creation is labor-intensive and time-consuming. Recent advancements of vision-language models (VLMs) have shown strong potential in medical applications, presenting a promising avenue for automating report generation. However, existing applications of VLMs in the medical domain have predominantly focused on structural imaging modalities, while the unique characteristics of molecular PET imaging have largely been overlooked. To bridge the gap, we introduce PET2Rep, a large-scale comprehensive benchmark for evaluation of general and medical VLMs for radiology report generation for PET images. PET2Rep stands out as the first dedicated dataset for PET report generation with metabolic information, uniquely capturing whole-body image-report pairs that cover dozens of organs to fill the critical gap in existing benchmarks and mirror real-world clinical comprehensiveness. In addition to widely recognized natural language generation metrics, we introduce a series of clinical efficiency metrics to evaluate the quality of radiotracer uptake pattern description in key organs in generated reports. We conduct a head-to-head comparison of 30 cutting-edge general-purpose and medical-specialized VLMs. The results show that the current state-of-the-art VLMs perform poorly on PET report generation task, falling considerably short of fulfilling practical needs. Moreover, we identify several key insufficiency that need to be addressed to advance the development in medical applications.
PAST: A multimodal single-cell foundation model for histopathology and spatial transcriptomics in cancer
Jul 08, 2025Abstract:While pathology foundation models have transformed cancer image analysis, they often lack integration with molecular data at single-cell resolution, limiting their utility for precision oncology. Here, we present PAST, a pan-cancer single-cell foundation model trained on 20 million paired histopathology images and single-cell transcriptomes spanning multiple tumor types and tissue contexts. By jointly encoding cellular morphology and gene expression, PAST learns unified cross-modal representations that capture both spatial and molecular heterogeneity at the cellular level. This approach enables accurate prediction of single-cell gene expression, virtual molecular staining, and multimodal survival analysis directly from routine pathology slides. Across diverse cancers and downstream tasks, PAST consistently exceeds the performance of existing approaches, demonstrating robust generalizability and scalability. Our work establishes a new paradigm for pathology foundation models, providing a versatile tool for high-resolution spatial omics, mechanistic discovery, and precision cancer research.
Structure-aware Semantic Discrepancy and Consistency for 3D Medical Image Self-supervised Learning
Jul 03, 2025Abstract:3D medical image self-supervised learning (mSSL) holds great promise for medical analysis. Effectively supporting broader applications requires considering anatomical structure variations in location, scale, and morphology, which are crucial for capturing meaningful distinctions. However, previous mSSL methods partition images with fixed-size patches, often ignoring the structure variations. In this work, we introduce a novel perspective on 3D medical images with the goal of learning structure-aware representations. We assume that patches within the same structure share the same semantics (semantic consistency) while those from different structures exhibit distinct semantics (semantic discrepancy). Based on this assumption, we propose an mSSL framework named $S^2DC$, achieving Structure-aware Semantic Discrepancy and Consistency in two steps. First, $S^2DC$ enforces distinct representations for different patches to increase semantic discrepancy by leveraging an optimal transport strategy. Second, $S^2DC$ advances semantic consistency at the structural level based on neighborhood similarity distribution. By bridging patch-level and structure-level representations, $S^2DC$ achieves structure-aware representations. Thoroughly evaluated across 10 datasets, 4 tasks, and 3 modalities, our proposed method consistently outperforms the state-of-the-art methods in mSSL.
Efficient Network Automatic Relevance Determination
Jun 14, 2025



Abstract:We propose Network Automatic Relevance Determination (NARD), an extension of ARD for linearly probabilistic models, to simultaneously model sparse relationships between inputs $X \in \mathbb R^{d \times N}$ and outputs $Y \in \mathbb R^{m \times N}$, while capturing the correlation structure among the $Y$. NARD employs a matrix normal prior which contains a sparsity-inducing parameter to identify and discard irrelevant features, thereby promoting sparsity in the model. Algorithmically, it iteratively updates both the precision matrix and the relationship between $Y$ and the refined inputs. To mitigate the computational inefficiencies of the $\mathcal O(m^3 + d^3)$ cost per iteration, we introduce Sequential NARD, which evaluates features sequentially, and a Surrogate Function Method, leveraging an efficient approximation of the marginal likelihood and simplifying the calculation of determinant and inverse of an intermediate matrix. Combining the Sequential update with the Surrogate Function method further reduces computational costs. The computational complexity per iteration for these three methods is reduced to $\mathcal O(m^3+p^3)$, $\mathcal O(m^3 + d^2)$, $\mathcal O(m^3+p^2)$, respectively, where $p \ll d$ is the final number of features in the model. Our methods demonstrate significant improvements in computational efficiency with comparable performance on both synthetic and real-world datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge