Zixin Hu
FSX: Message Flow Sensitivity Enhanced Structural Explainer for Graph Neural Networks
Jan 21, 2026Abstract:Despite the widespread success of Graph Neural Networks (GNNs), understanding the reasons behind their specific predictions remains challenging. Existing explainability methods face a trade-off that gradient-based approaches are computationally efficient but often ignore structural interactions, while game-theoretic techniques capture interactions at the cost of high computational overhead and potential deviation from the model's true reasoning path. To address this gap, we propose FSX (Message Flow Sensitivity Enhanced Structural Explainer), a novel hybrid framework that synergistically combines the internal message flows of the model with a cooperative game approach applied to the external graph data. FSX first identifies critical message flows via a novel flow-sensitivity analysis: during a single forward pass, it simulates localized node perturbations and measures the resulting changes in message flow intensities. These sensitivity-ranked flows are then projected onto the input graph to define compact, semantically meaningful subgraphs. Within each subgraph, a flow-aware cooperative game is conducted, where node contributions are evaluated fairly through a Shapley-like value that incorporates both node-feature importance and their roles in sustaining or destabilizing the identified critical flows. Extensive evaluation across multiple datasets and GNN architectures demonstrates that FSX achieves superior explanation fidelity with significantly reduced runtime, while providing unprecedented insights into the structural logic underlying model predictions--specifically, how important sub-structures exert influence by governing the stability of key internal computational pathways.
PET2Rep: Towards Vision-Language Model-Drived Automated Radiology Report Generation for Positron Emission Tomography
Aug 06, 2025Abstract:Positron emission tomography (PET) is a cornerstone of modern oncologic and neurologic imaging, distinguished by its unique ability to illuminate dynamic metabolic processes that transcend the anatomical focus of traditional imaging technologies. Radiology reports are essential for clinical decision making, yet their manual creation is labor-intensive and time-consuming. Recent advancements of vision-language models (VLMs) have shown strong potential in medical applications, presenting a promising avenue for automating report generation. However, existing applications of VLMs in the medical domain have predominantly focused on structural imaging modalities, while the unique characteristics of molecular PET imaging have largely been overlooked. To bridge the gap, we introduce PET2Rep, a large-scale comprehensive benchmark for evaluation of general and medical VLMs for radiology report generation for PET images. PET2Rep stands out as the first dedicated dataset for PET report generation with metabolic information, uniquely capturing whole-body image-report pairs that cover dozens of organs to fill the critical gap in existing benchmarks and mirror real-world clinical comprehensiveness. In addition to widely recognized natural language generation metrics, we introduce a series of clinical efficiency metrics to evaluate the quality of radiotracer uptake pattern description in key organs in generated reports. We conduct a head-to-head comparison of 30 cutting-edge general-purpose and medical-specialized VLMs. The results show that the current state-of-the-art VLMs perform poorly on PET report generation task, falling considerably short of fulfilling practical needs. Moreover, we identify several key insufficiency that need to be addressed to advance the development in medical applications.
Efficient Network Automatic Relevance Determination
Jun 14, 2025



Abstract:We propose Network Automatic Relevance Determination (NARD), an extension of ARD for linearly probabilistic models, to simultaneously model sparse relationships between inputs $X \in \mathbb R^{d \times N}$ and outputs $Y \in \mathbb R^{m \times N}$, while capturing the correlation structure among the $Y$. NARD employs a matrix normal prior which contains a sparsity-inducing parameter to identify and discard irrelevant features, thereby promoting sparsity in the model. Algorithmically, it iteratively updates both the precision matrix and the relationship between $Y$ and the refined inputs. To mitigate the computational inefficiencies of the $\mathcal O(m^3 + d^3)$ cost per iteration, we introduce Sequential NARD, which evaluates features sequentially, and a Surrogate Function Method, leveraging an efficient approximation of the marginal likelihood and simplifying the calculation of determinant and inverse of an intermediate matrix. Combining the Sequential update with the Surrogate Function method further reduces computational costs. The computational complexity per iteration for these three methods is reduced to $\mathcal O(m^3+p^3)$, $\mathcal O(m^3 + d^2)$, $\mathcal O(m^3+p^2)$, respectively, where $p \ll d$ is the final number of features in the model. Our methods demonstrate significant improvements in computational efficiency with comparable performance on both synthetic and real-world datasets.
FuXi-S2S: An accurate machine learning model for global subseasonal forecasts
Dec 15, 2023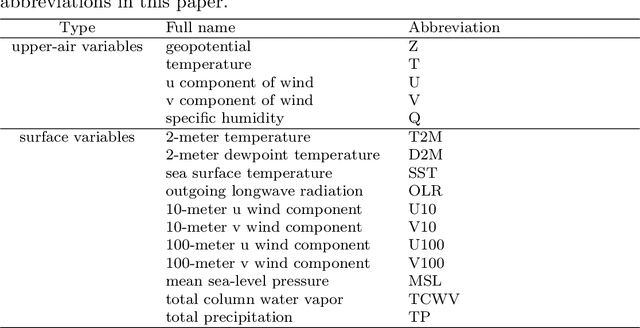
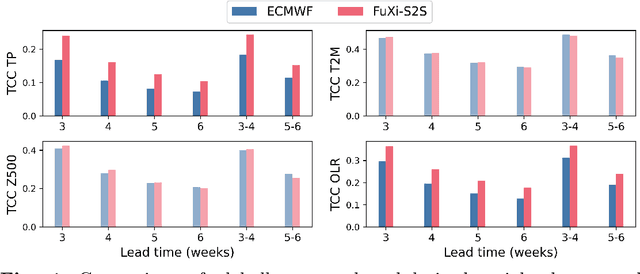
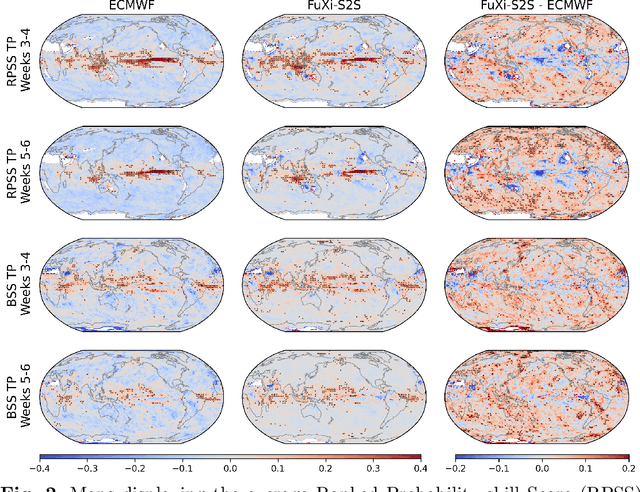
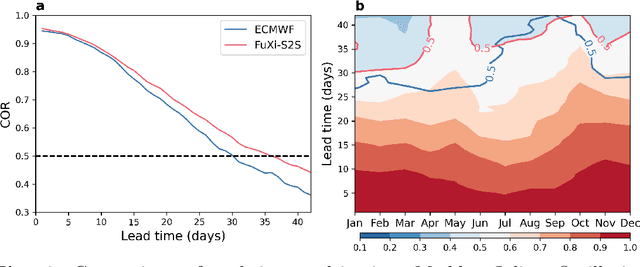
Abstract:Skillful subseasonal forecasts beyond 2 weeks are crucial for a wide range of applications across various sectors of society. Recently, state-of-the-art machine learning based weather forecasting models have made significant advancements, outperforming the high-resolution forecast (HRES) from the European Centre for Medium-Range Weather Forecasts (ECMWF). However, the full potential of machine learning models in subseasonal forecasts has yet to be fully explored. In this study, we introduce FuXi Subseasonal-to-Seasonal (FuXi-S2S), a machine learning based subseasonal forecasting model that provides global daily mean forecasts up to 42 days, covering 5 upper-air atmospheric variables at 13 pressure levels and 11 surface variables. FuXi-S2S integrates an enhanced FuXi base model with a perturbation module for flow-dependent perturbations in hidden features, and incorporates Perlin noise to perturb initial conditions. The model is developed using 72 years of daily statistics from ECMWF ERA5 reanalysis data. When compared to the ECMWF Subseasonal-to-Seasonal (S2S) reforecasts, the FuXi-S2S forecasts demonstrate superior deterministic and ensemble forecasts for total precipitation (TP), outgoing longwave radiation (OLR), and geopotential at 500 hPa (Z500). Although it shows slightly inferior performance in predicting 2-meter temperature (T2M), it has clear advantages over land area. Regarding the extreme forecasts, FuXi-S2S outperforms ECMWF S2S globally for TP. Furthermore, FuXi-S2S forecasts surpass the ECMWF S2S reforecasts in predicting the Madden Julian Oscillation (MJO), a key source of subseasonal predictability. They extend the skillful prediction of MJO from 30 days to 36 days.
CoGANPPIS: Coevolution-enhanced Global Attention Neural Network for Protein-Protein Interaction Site Prediction
Apr 03, 2023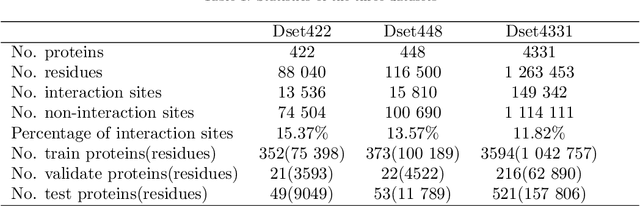
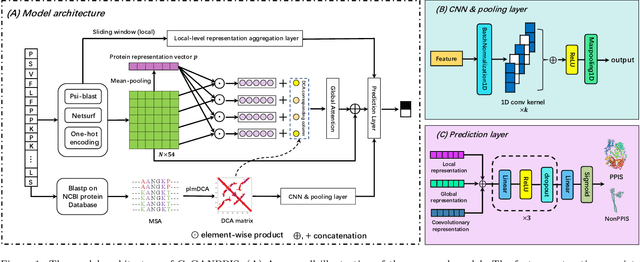
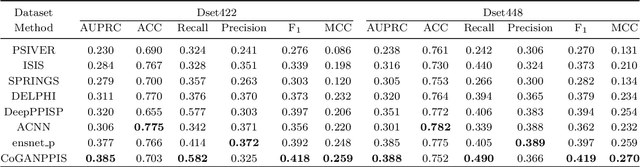
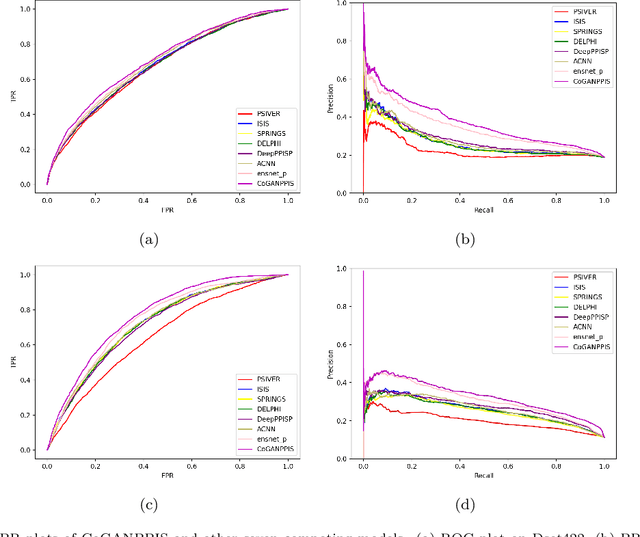
Abstract:Protein-protein interactions are essential in biochemical processes. Accurate prediction of the protein-protein interaction sites (PPIs) deepens our understanding of biological mechanism and is crucial for new drug design. However, conventional experimental methods for PPIs prediction are costly and time-consuming so that many computational approaches, especially ML-based methods, have been developed recently. Although these approaches have achieved gratifying results, there are still two limitations: (1) Most models have excavated some useful input features, but failed to take coevolutionary features into account, which could provide clues for inter-residue relationships; (2) The attention-based models only allocate attention weights for neighboring residues, instead of doing it globally, neglecting that some residues being far away from the target residues might also matter. We propose a coevolution-enhanced global attention neural network, a sequence-based deep learning model for PPIs prediction, called CoGANPPIS. It utilizes three layers in parallel for feature extraction: (1) Local-level representation aggregation layer, which aggregates the neighboring residues' features; (2) Global-level representation learning layer, which employs a novel coevolution-enhanced global attention mechanism to allocate attention weights to all the residues on the same protein sequences; (3) Coevolutionary information learning layer, which applies CNN & pooling to coevolutionary information to obtain the coevolutionary profile representation. Then, the three outputs are concatenated and passed into several fully connected layers for the final prediction. Application on two benchmark datasets demonstrated a state-of-the-art performance of our model. The source code is publicly available at https://github.com/Slam1423/CoGANPPIS_source_code.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge