Xiaoxi Hu
Recurrent Cross-View Object Geo-Localization
Sep 16, 2025Abstract:Cross-view object geo-localization (CVOGL) aims to determine the location of a specific object in high-resolution satellite imagery given a query image with a point prompt. Existing approaches treat CVOGL as a one-shot detection task, directly regressing object locations from cross-view information aggregation, but they are vulnerable to feature noise and lack mechanisms for error correction. In this paper, we propose ReCOT, a Recurrent Cross-view Object geo-localization Transformer, which reformulates CVOGL as a recurrent localization task. ReCOT introduces a set of learnable tokens that encode task-specific intent from the query image and prompt embeddings, and iteratively attend to the reference features to refine the predicted location. To enhance this recurrent process, we incorporate two complementary modules: (1) a SAM-based knowledge distillation strategy that transfers segmentation priors from the Segment Anything Model (SAM) to provide clearer semantic guidance without additional inference cost, and (2) a Reference Feature Enhancement Module (RFEM) that introduces a hierarchical attention to emphasize object-relevant regions in the reference features. Extensive experiments on standard CVOGL benchmarks demonstrate that ReCOT achieves state-of-the-art (SOTA) performance while reducing parameters by 60% compared to previous SOTA approaches.
Progressive Bird's Eye View Perception for Safety-Critical Autonomous Driving: A Comprehensive Survey
Aug 11, 2025Abstract:Bird's-Eye-View (BEV) perception has become a foundational paradigm in autonomous driving, enabling unified spatial representations that support robust multi-sensor fusion and multi-agent collaboration. As autonomous vehicles transition from controlled environments to real-world deployment, ensuring the safety and reliability of BEV perception in complex scenarios - such as occlusions, adverse weather, and dynamic traffic - remains a critical challenge. This survey provides the first comprehensive review of BEV perception from a safety-critical perspective, systematically analyzing state-of-the-art frameworks and implementation strategies across three progressive stages: single-modality vehicle-side, multimodal vehicle-side, and multi-agent collaborative perception. Furthermore, we examine public datasets encompassing vehicle-side, roadside, and collaborative settings, evaluating their relevance to safety and robustness. We also identify key open-world challenges - including open-set recognition, large-scale unlabeled data, sensor degradation, and inter-agent communication latency - and outline future research directions, such as integration with end-to-end autonomous driving systems, embodied intelligence, and large language models.
Collaborative Perception Datasets for Autonomous Driving: A Review
Apr 17, 2025



Abstract:Collaborative perception has attracted growing interest from academia and industry due to its potential to enhance perception accuracy, safety, and robustness in autonomous driving through multi-agent information fusion. With the advancement of Vehicle-to-Everything (V2X) communication, numerous collaborative perception datasets have emerged, varying in cooperation paradigms, sensor configurations, data sources, and application scenarios. However, the absence of systematic summarization and comparative analysis hinders effective resource utilization and standardization of model evaluation. As the first comprehensive review focused on collaborative perception datasets, this work reviews and compares existing resources from a multi-dimensional perspective. We categorize datasets based on cooperation paradigms, examine their data sources and scenarios, and analyze sensor modalities and supported tasks. A detailed comparative analysis is conducted across multiple dimensions. We also outline key challenges and future directions, including dataset scalability, diversity, domain adaptation, standardization, privacy, and the integration of large language models. To support ongoing research, we provide a continuously updated online repository of collaborative perception datasets and related literature: https://github.com/frankwnb/Collaborative-Perception-Datasets-for-Autonomous-Driving.
CoGANPPIS: Coevolution-enhanced Global Attention Neural Network for Protein-Protein Interaction Site Prediction
Apr 03, 2023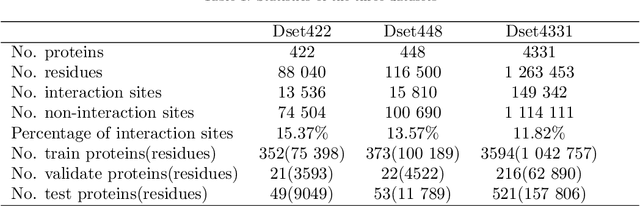
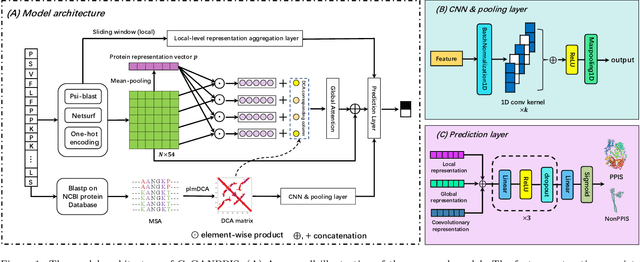
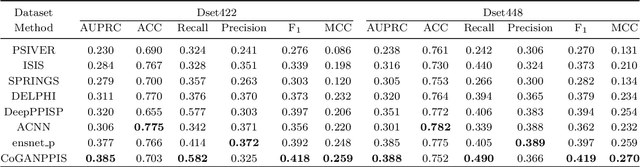
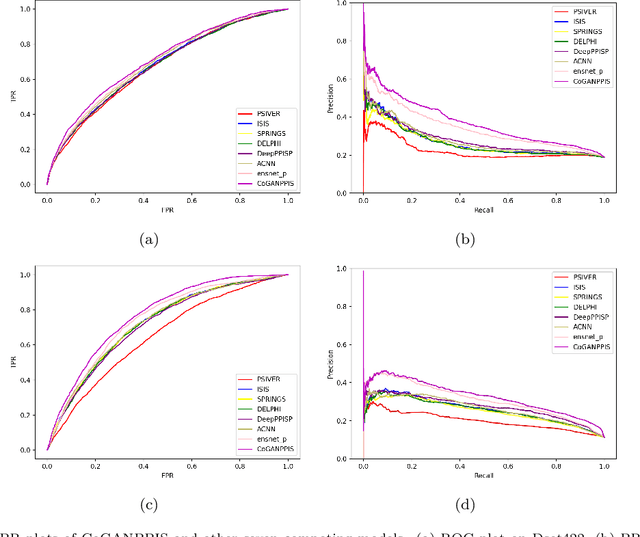
Abstract:Protein-protein interactions are essential in biochemical processes. Accurate prediction of the protein-protein interaction sites (PPIs) deepens our understanding of biological mechanism and is crucial for new drug design. However, conventional experimental methods for PPIs prediction are costly and time-consuming so that many computational approaches, especially ML-based methods, have been developed recently. Although these approaches have achieved gratifying results, there are still two limitations: (1) Most models have excavated some useful input features, but failed to take coevolutionary features into account, which could provide clues for inter-residue relationships; (2) The attention-based models only allocate attention weights for neighboring residues, instead of doing it globally, neglecting that some residues being far away from the target residues might also matter. We propose a coevolution-enhanced global attention neural network, a sequence-based deep learning model for PPIs prediction, called CoGANPPIS. It utilizes three layers in parallel for feature extraction: (1) Local-level representation aggregation layer, which aggregates the neighboring residues' features; (2) Global-level representation learning layer, which employs a novel coevolution-enhanced global attention mechanism to allocate attention weights to all the residues on the same protein sequences; (3) Coevolutionary information learning layer, which applies CNN & pooling to coevolutionary information to obtain the coevolutionary profile representation. Then, the three outputs are concatenated and passed into several fully connected layers for the final prediction. Application on two benchmark datasets demonstrated a state-of-the-art performance of our model. The source code is publicly available at https://github.com/Slam1423/CoGANPPIS_source_code.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge