Fenglei Cao
A Pre-trained Reaction Embedding Descriptor Capturing Bond Transformation Patterns
Jan 07, 2026Abstract:With the rise of data-driven reaction prediction models, effective reaction descriptors are crucial for bridging the gap between real-world chemistry and digital representations. However, general-purpose, reaction-wise descriptors remain scarce. This study introduces RXNEmb, a novel reaction-level descriptor derived from RXNGraphormer, a model pre-trained to distinguish real reactions from fictitious ones with erroneous bond changes, thereby learning intrinsic bond formation and cleavage patterns. We demonstrate its utility by data-driven re-clustering of the USPTO-50k dataset, yielding a classification that more directly reflects bond-change similarities than rule-based categories. Combined with dimensionality reduction, RXNEmb enables visualization of reaction space diversity. Furthermore, attention weight analysis reveals the model's focus on chemically critical sites, providing mechanistic insight. RXNEmb serves as a powerful, interpretable tool for reaction fingerprinting and analysis, paving the way for more data-centric approaches in reaction analysis and discovery.
Equivariant Spherical Transformer for Efficient Molecular Modeling
May 29, 2025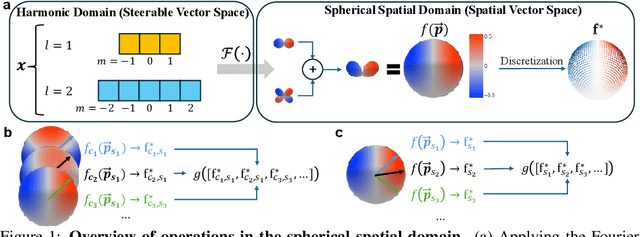
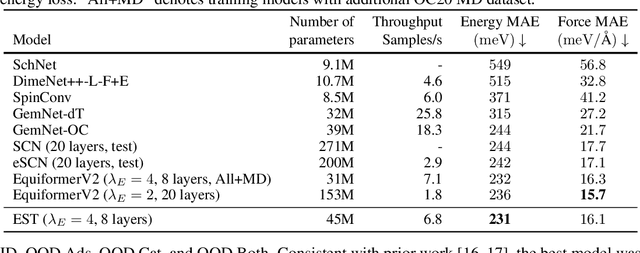
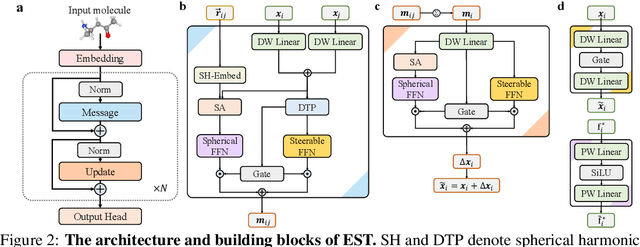
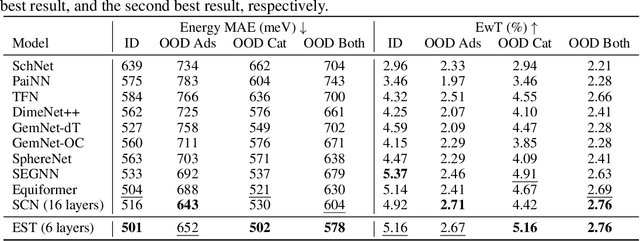
Abstract:SE(3)-equivariant Graph Neural Networks (GNNs) have significantly advanced molecular system modeling by employing group representations. However, their message passing processes, which rely on tensor product-based convolutions, are limited by insufficient non-linearity and incomplete group representations, thereby restricting expressiveness. To overcome these limitations, we introduce the Equivariant Spherical Transformer (EST), a novel framework that leverages a Transformer structure within the spatial domain of group representations after Fourier transform. We theoretically and empirically demonstrate that EST can encompass the function space of tensor products while achieving superior expressiveness. Furthermore, EST's equivariant inductive bias is guaranteed through a uniform sampling strategy for the Fourier transform. Our experiments demonstrate state-of-the-art performance by EST on various molecular benchmarks, including OC20 and QM9.
ChemHAS: Hierarchical Agent Stacking for Enhancing Chemistry Tools
May 27, 2025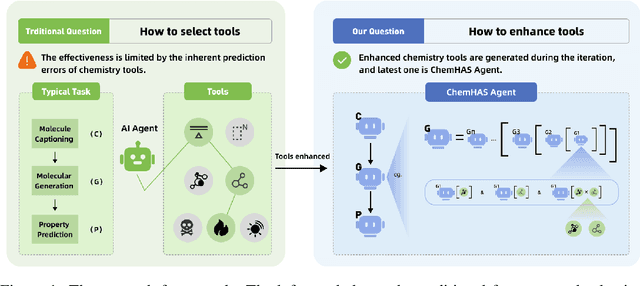
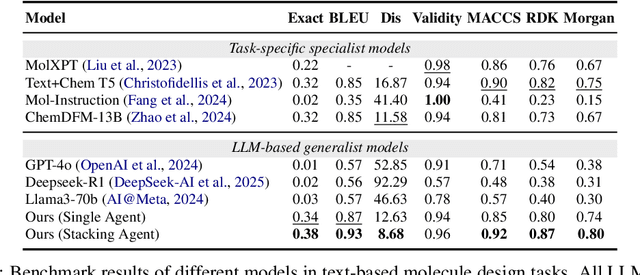
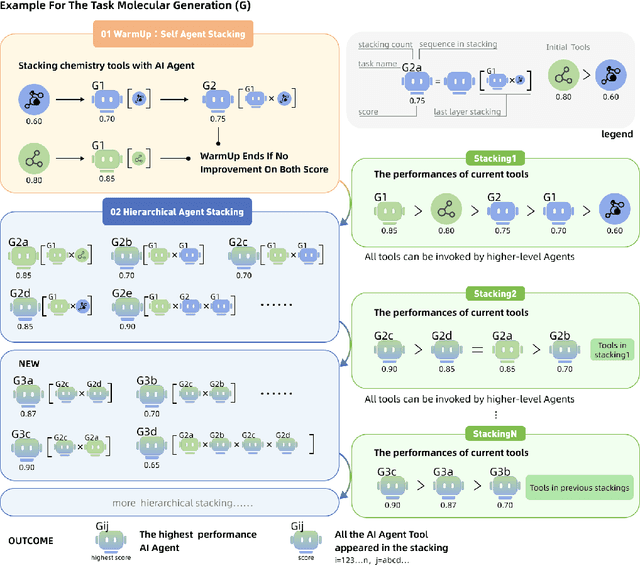
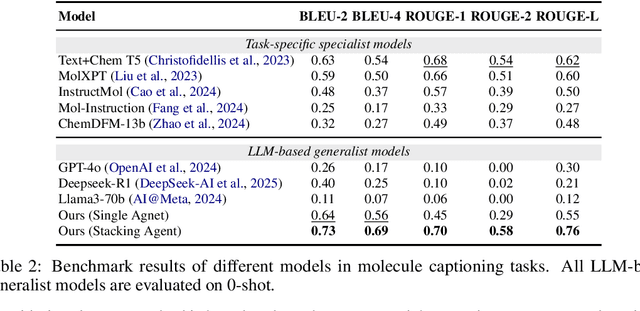
Abstract:Large Language Model (LLM)-based agents have demonstrated the ability to improve performance in chemistry-related tasks by selecting appropriate tools. However, their effectiveness remains limited by the inherent prediction errors of chemistry tools. In this paper, we take a step further by exploring how LLMbased agents can, in turn, be leveraged to reduce prediction errors of the tools. To this end, we propose ChemHAS (Chemical Hierarchical Agent Stacking), a simple yet effective method that enhances chemistry tools through optimizing agent-stacking structures from limited data. ChemHAS achieves state-of-the-art performance across four fundamental chemistry tasks, demonstrating that our method can effectively compensate for prediction errors of the tools. Furthermore, we identify and characterize four distinct agent-stacking behaviors, potentially improving interpretability and revealing new possibilities for AI agent applications in scientific research. Our code and dataset are publicly available at https: //anonymous.4open.science/r/ChemHAS-01E4/README.md.
Equivariant Masked Position Prediction for Efficient Molecular Representation
Feb 12, 2025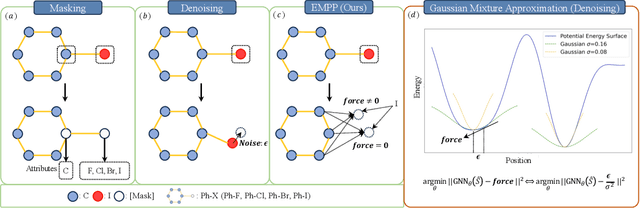
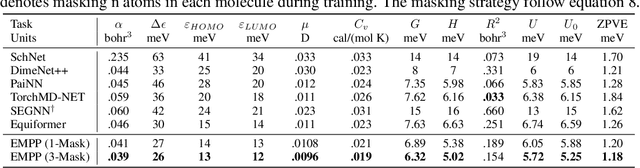
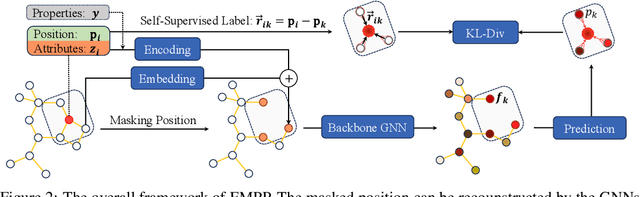
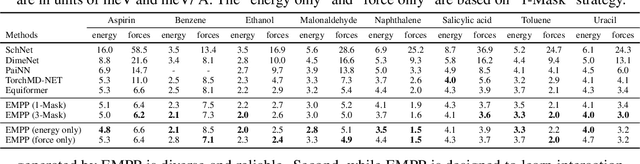
Abstract:Graph neural networks (GNNs) have shown considerable promise in computational chemistry. However, the limited availability of molecular data raises concerns regarding GNNs' ability to effectively capture the fundamental principles of physics and chemistry, which constrains their generalization capabilities. To address this challenge, we introduce a novel self-supervised approach termed Equivariant Masked Position Prediction (EMPP), grounded in intramolecular potential and force theory. Unlike conventional attribute masking techniques, EMPP formulates a nuanced position prediction task that is more well-defined and enhances the learning of quantum mechanical features. EMPP also bypasses the approximation of the Gaussian mixture distribution commonly used in denoising methods, allowing for more accurate acquisition of physical properties. Experimental results indicate that EMPP significantly enhances performance of advanced molecular architectures, surpassing state-of-the-art self-supervised approaches. Our code is released in https://github.com/ajy112/EMPP.
* 24 pages, 6 figures
Dynamic PDB: A New Dataset and a SE(3) Model Extension by Integrating Dynamic Behaviors and Physical Properties in Protein Structures
Aug 22, 2024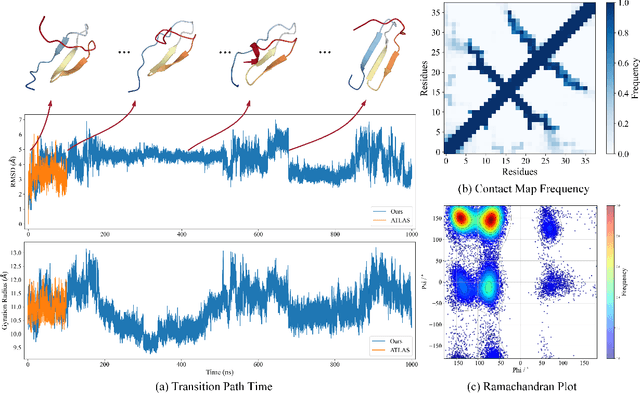
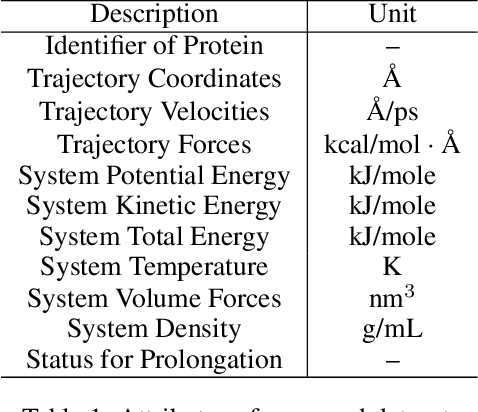
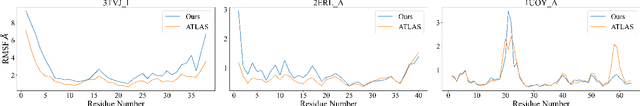
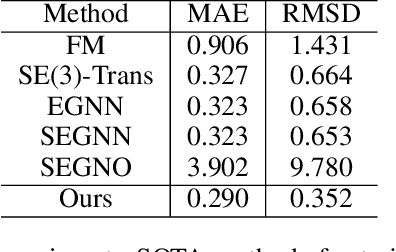
Abstract:Despite significant progress in static protein structure collection and prediction, the dynamic behavior of proteins, one of their most vital characteristics, has been largely overlooked in prior research. This oversight can be attributed to the limited availability, diversity, and heterogeneity of dynamic protein datasets. To address this gap, we propose to enhance existing prestigious static 3D protein structural databases, such as the Protein Data Bank (PDB), by integrating dynamic data and additional physical properties. Specifically, we introduce a large-scale dataset, Dynamic PDB, encompassing approximately 12.6K proteins, each subjected to all-atom molecular dynamics (MD) simulations lasting 1 microsecond to capture conformational changes. Furthermore, we provide a comprehensive suite of physical properties, including atomic velocities and forces, potential and kinetic energies of proteins, and the temperature of the simulation environment, recorded at 1 picosecond intervals throughout the simulations. For benchmarking purposes, we evaluate state-of-the-art methods on the proposed dataset for the task of trajectory prediction. To demonstrate the value of integrating richer physical properties in the study of protein dynamics and related model design, we base our approach on the SE(3) diffusion model and incorporate these physical properties into the trajectory prediction process. Preliminary results indicate that this straightforward extension of the SE(3) model yields improved accuracy, as measured by MAE and RMSD, when the proposed physical properties are taken into consideration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge