Liwei Zhang
Generalize Your Face Forgery Detectors: An Insertable Adaptation Module Is All You Need
Dec 30, 2024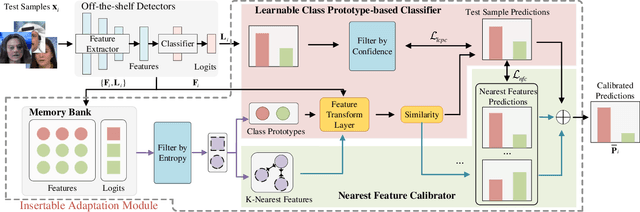
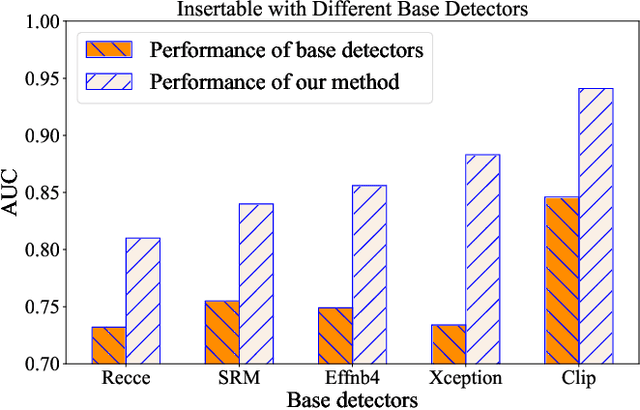
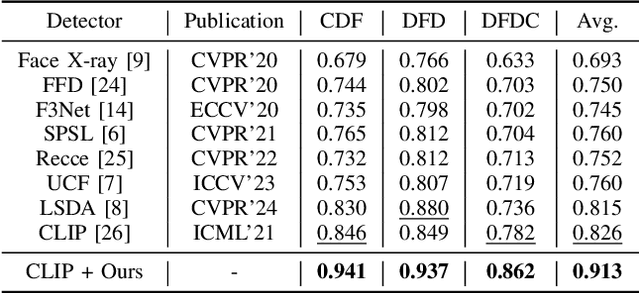

Abstract:A plethora of face forgery detectors exist to tackle facial deepfake risks. However, their practical application is hindered by the challenge of generalizing to forgeries unseen during the training stage. To this end, we introduce an insertable adaptation module that can adapt a trained off-the-shelf detector using only online unlabeled test data, without requiring modifications to the architecture or training process. Specifically, we first present a learnable class prototype-based classifier that generates predictions from the revised features and prototypes, enabling effective handling of various forgery clues and domain gaps during online testing. Additionally, we propose a nearest feature calibrator to further improve prediction accuracy and reduce the impact of noisy pseudo-labels during self-training. Experiments across multiple datasets show that our module achieves superior generalization compared to state-of-the-art methods. Moreover, it functions as a plug-and-play component that can be combined with various detectors to enhance the overall performance.
Dynamic PDB: A New Dataset and a SE(3) Model Extension by Integrating Dynamic Behaviors and Physical Properties in Protein Structures
Aug 22, 2024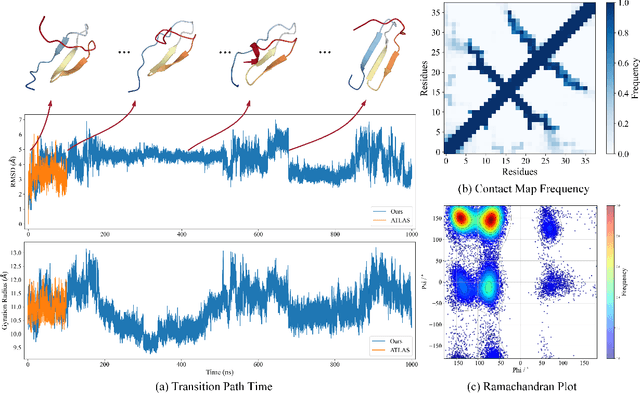
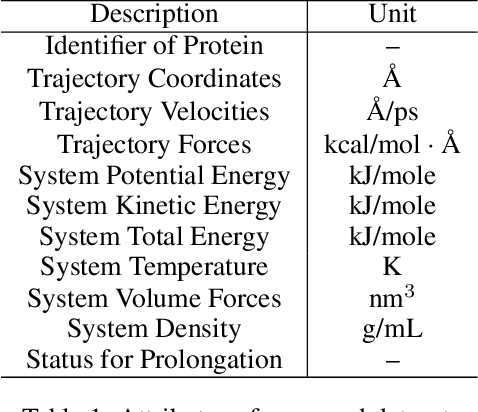
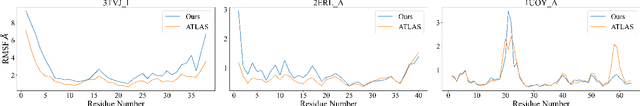
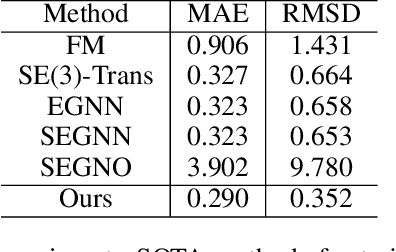
Abstract:Despite significant progress in static protein structure collection and prediction, the dynamic behavior of proteins, one of their most vital characteristics, has been largely overlooked in prior research. This oversight can be attributed to the limited availability, diversity, and heterogeneity of dynamic protein datasets. To address this gap, we propose to enhance existing prestigious static 3D protein structural databases, such as the Protein Data Bank (PDB), by integrating dynamic data and additional physical properties. Specifically, we introduce a large-scale dataset, Dynamic PDB, encompassing approximately 12.6K proteins, each subjected to all-atom molecular dynamics (MD) simulations lasting 1 microsecond to capture conformational changes. Furthermore, we provide a comprehensive suite of physical properties, including atomic velocities and forces, potential and kinetic energies of proteins, and the temperature of the simulation environment, recorded at 1 picosecond intervals throughout the simulations. For benchmarking purposes, we evaluate state-of-the-art methods on the proposed dataset for the task of trajectory prediction. To demonstrate the value of integrating richer physical properties in the study of protein dynamics and related model design, we base our approach on the SE(3) diffusion model and incorporate these physical properties into the trajectory prediction process. Preliminary results indicate that this straightforward extension of the SE(3) model yields improved accuracy, as measured by MAE and RMSD, when the proposed physical properties are taken into consideration.
4D Diffusion for Dynamic Protein Structure Prediction with Reference Guided Motion Alignment
Aug 22, 2024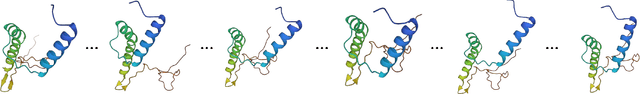
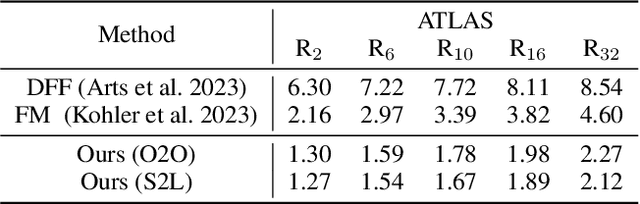
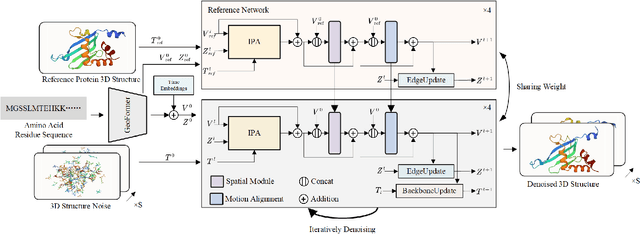
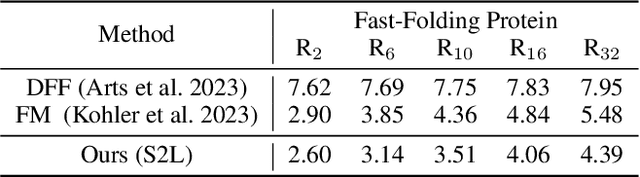
Abstract:Protein structure prediction is pivotal for understanding the structure-function relationship of proteins, advancing biological research, and facilitating pharmaceutical development and experimental design. While deep learning methods and the expanded availability of experimental 3D protein structures have accelerated structure prediction, the dynamic nature of protein structures has received limited attention. This study introduces an innovative 4D diffusion model incorporating molecular dynamics (MD) simulation data to learn dynamic protein structures. Our approach is distinguished by the following components: (1) a unified diffusion model capable of generating dynamic protein structures, including both the backbone and side chains, utilizing atomic grouping and side-chain dihedral angle predictions; (2) a reference network that enhances structural consistency by integrating the latent embeddings of the initial 3D protein structures; and (3) a motion alignment module aimed at improving temporal structural coherence across multiple time steps. To our knowledge, this is the first diffusion-based model aimed at predicting protein trajectories across multiple time steps simultaneously. Validation on benchmark datasets demonstrates that our model exhibits high accuracy in predicting dynamic 3D structures of proteins containing up to 256 amino acids over 32 time steps, effectively capturing both local flexibility in stable states and significant conformational changes.
Hallo: Hierarchical Audio-Driven Visual Synthesis for Portrait Image Animation
Jun 16, 2024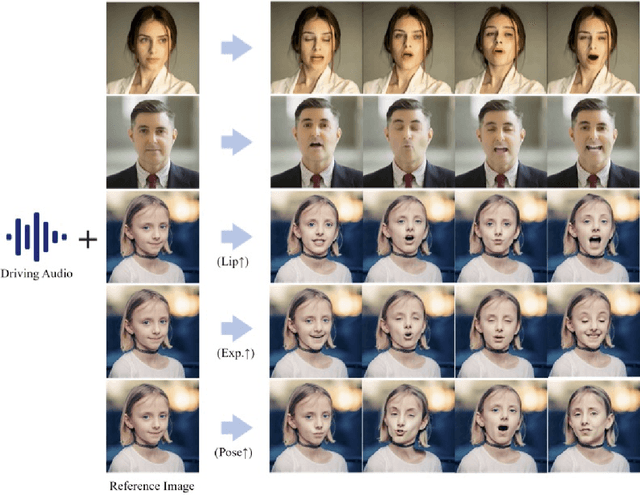
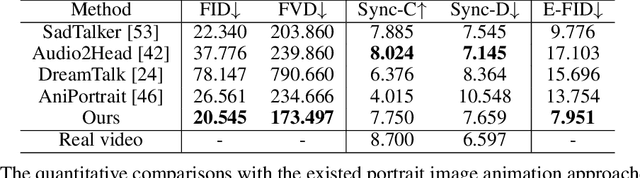
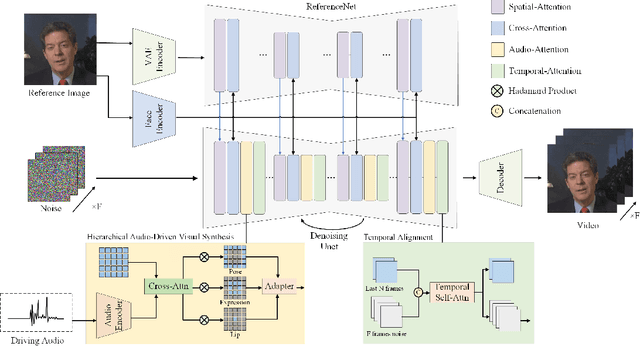
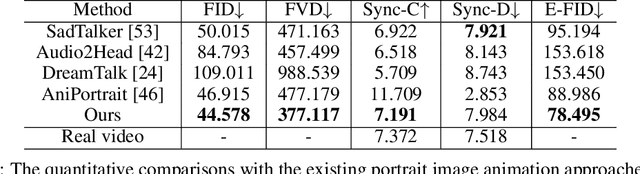
Abstract:The field of portrait image animation, driven by speech audio input, has experienced significant advancements in the generation of realistic and dynamic portraits. This research delves into the complexities of synchronizing facial movements and creating visually appealing, temporally consistent animations within the framework of diffusion-based methodologies. Moving away from traditional paradigms that rely on parametric models for intermediate facial representations, our innovative approach embraces the end-to-end diffusion paradigm and introduces a hierarchical audio-driven visual synthesis module to enhance the precision of alignment between audio inputs and visual outputs, encompassing lip, expression, and pose motion. Our proposed network architecture seamlessly integrates diffusion-based generative models, a UNet-based denoiser, temporal alignment techniques, and a reference network. The proposed hierarchical audio-driven visual synthesis offers adaptive control over expression and pose diversity, enabling more effective personalization tailored to different identities. Through a comprehensive evaluation that incorporates both qualitative and quantitative analyses, our approach demonstrates obvious enhancements in image and video quality, lip synchronization precision, and motion diversity. Further visualization and access to the source code can be found at: https://fudan-generative-vision.github.io/hallo.
Augmented Lagrangian Methods for Time-varying Constrained Online Convex Optimization
May 19, 2022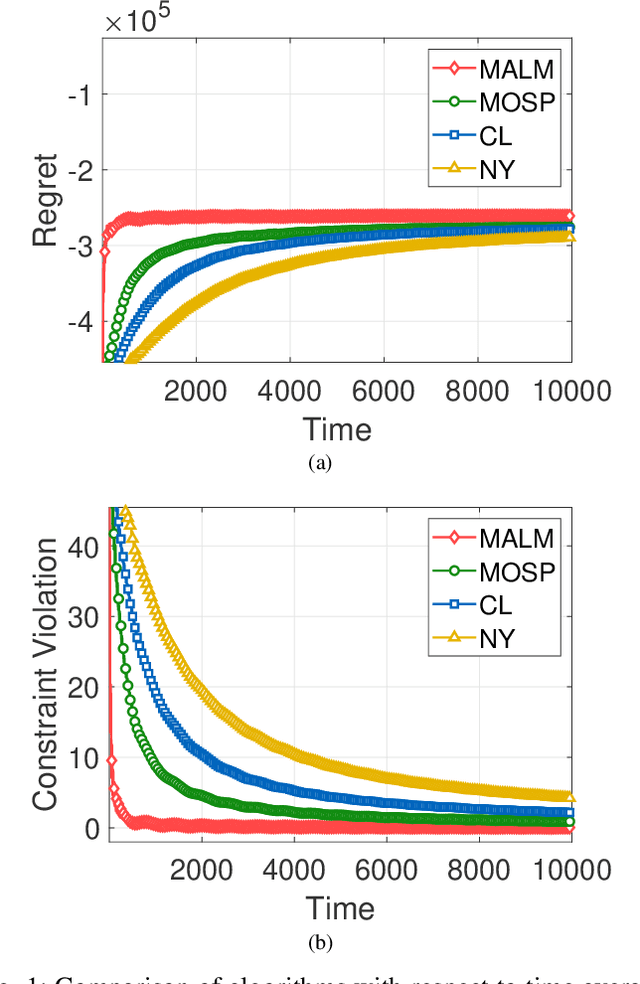

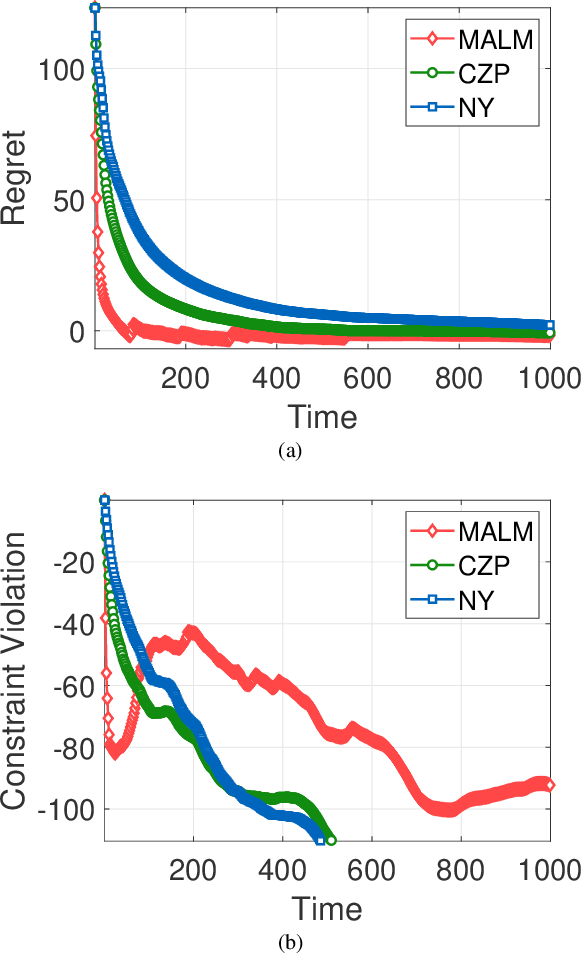
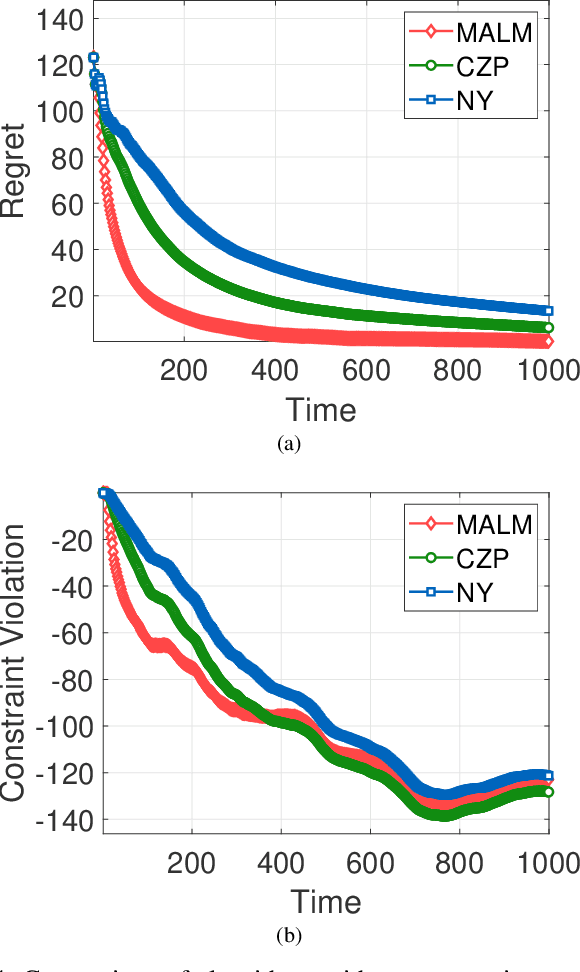
Abstract:In this paper, we consider online convex optimization (OCO) with time-varying loss and constraint functions. Specifically, the decision maker chooses sequential decisions based only on past information, meantime the loss and constraint functions are revealed over time. We first develop a class of model-based augmented Lagrangian methods (MALM) for time-varying functional constrained OCO (without feedback delay). Under standard assumptions, we establish sublinear regret and sublinear constraint violation of MALM. Furthermore, we extend MALM to deal with time-varying functional constrained OCO with delayed feedback, in which the feedback information of loss and constraint functions is revealed to decision maker with delays. Without additional assumptions, we also establish sublinear regret and sublinear constraint violation for the delayed version of MALM. Finally, numerical results for several examples of constrained OCO including online network resource allocation, online logistic regression and online quadratically constrained quadratical program are presented to demonstrate the efficiency of the proposed algorithms.
A stochastic linearized proximal method of multipliers for convex stochastic optimization with expectation constraints
Jun 22, 2021

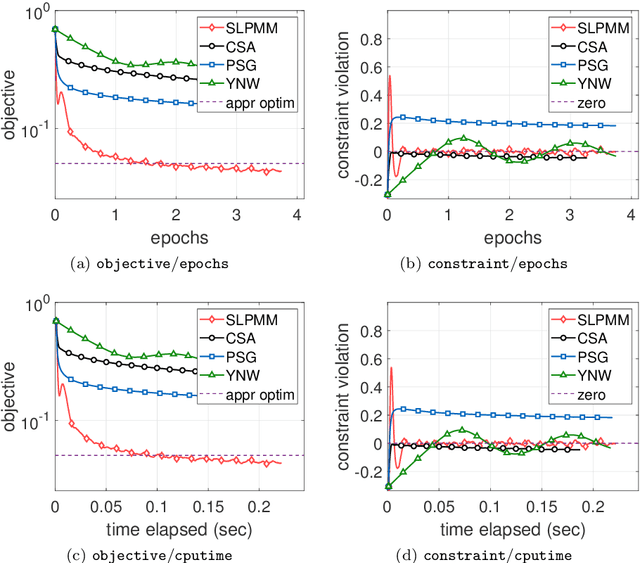

Abstract:This paper considers the problem of minimizing a convex expectation function with a set of inequality convex expectation constraints. We present a computable stochastic approximation type algorithm, namely the stochastic linearized proximal method of multipliers, to solve this convex stochastic optimization problem. This algorithm can be roughly viewed as a hybrid of stochastic approximation and the traditional proximal method of multipliers. Under mild conditions, we show that this algorithm exhibits $O(K^{-1/2})$ expected convergence rates for both objective reduction and constraint violation if parameters in the algorithm are properly chosen, where $K$ denotes the number of iterations. Moreover, we show that, with high probability, the algorithm has $O(\log(K)K^{-1/2})$ constraint violation bound and $O(\log^{3/2}(K)K^{-1/2})$ objective bound. Some preliminary numerical results demonstrate the performance of the proposed algorithm.
Synergistic Drug Combination Prediction by Integrating Multi-omics Data in Deep Learning Models
Nov 16, 2018
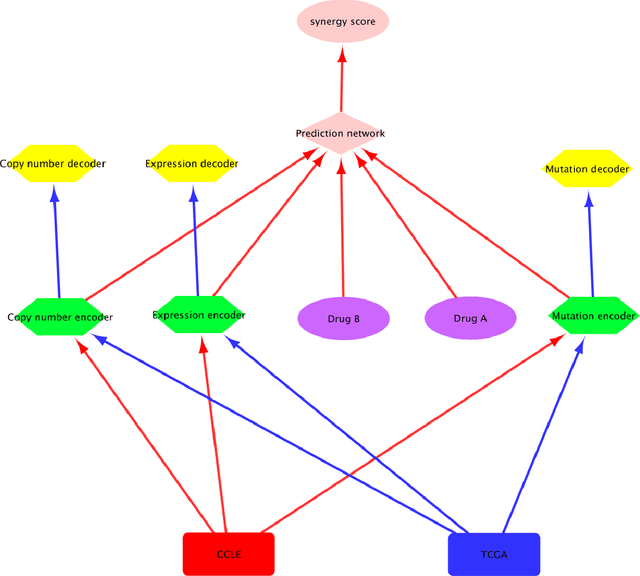
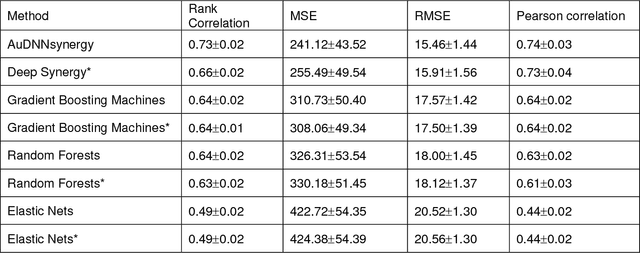
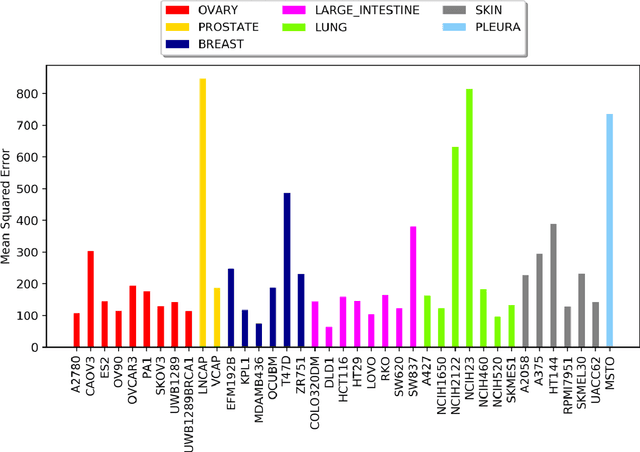
Abstract:Drug resistance is still a major challenge in cancer therapy. Drug combination is expected to overcome drug resistance. However, the number of possible drug combinations is enormous, and thus it is infeasible to experimentally screen all effective drug combinations considering the limited resources. Therefore, computational models to predict and prioritize effective drug combinations is important for combinatory therapy discovery in cancer. In this study, we proposed a novel deep learning model, AuDNNsynergy, to prediction drug combinations by integrating multi-omics data and chemical structure data. In specific, three autoencoders were trained using the gene expression, copy number and genetic mutation data of all tumor samples from The Cancer Genome Atlas. Then the physicochemical properties of drugs combined with the output of the three autoencoders, characterizing the individual cancer cell-lines, were used as the input of a deep neural network that predicts the synergy value of given pair-wise drug combinations against the specific cancer cell-lines. The comparison results showed the proposed AuDNNsynergy model outperforms four state-of-art approaches, namely DeepSynergy, Gradient Boosting Machines, Random Forests, and Elastic Nets. Moreover, we conducted the interpretation analysis of the deep learning model to investigate potential vital genetic predictors and the underlying mechanism of synergistic drug combinations on specific cancer cell-lines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge