Synergistic Drug Combination Prediction by Integrating Multi-omics Data in Deep Learning Models
Paper and Code
Nov 16, 2018
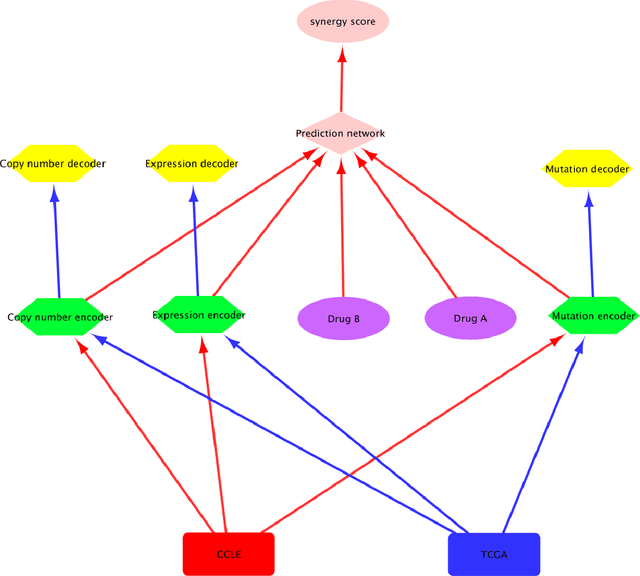
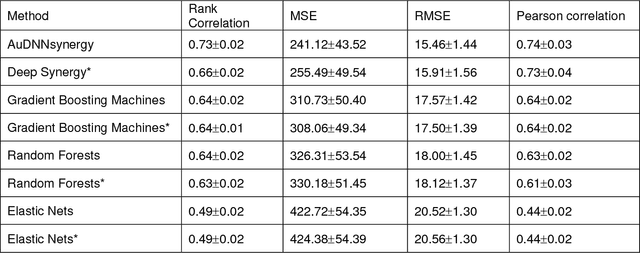
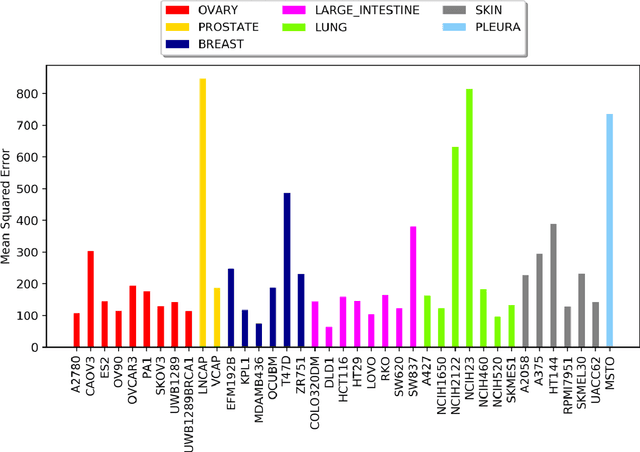
Drug resistance is still a major challenge in cancer therapy. Drug combination is expected to overcome drug resistance. However, the number of possible drug combinations is enormous, and thus it is infeasible to experimentally screen all effective drug combinations considering the limited resources. Therefore, computational models to predict and prioritize effective drug combinations is important for combinatory therapy discovery in cancer. In this study, we proposed a novel deep learning model, AuDNNsynergy, to prediction drug combinations by integrating multi-omics data and chemical structure data. In specific, three autoencoders were trained using the gene expression, copy number and genetic mutation data of all tumor samples from The Cancer Genome Atlas. Then the physicochemical properties of drugs combined with the output of the three autoencoders, characterizing the individual cancer cell-lines, were used as the input of a deep neural network that predicts the synergy value of given pair-wise drug combinations against the specific cancer cell-lines. The comparison results showed the proposed AuDNNsynergy model outperforms four state-of-art approaches, namely DeepSynergy, Gradient Boosting Machines, Random Forests, and Elastic Nets. Moreover, we conducted the interpretation analysis of the deep learning model to investigate potential vital genetic predictors and the underlying mechanism of synergistic drug combinations on specific cancer cell-lines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge