Philip R. O. Payne
Highly Accurate Disease Diagnosis and Highly Reproducible Biomarker Identification with PathFormer
Feb 11, 2024



Abstract:Biomarker identification is critical for precise disease diagnosis and understanding disease pathogenesis in omics data analysis, like using fold change and regression analysis. Graph neural networks (GNNs) have been the dominant deep learning model for analyzing graph-structured data. However, we found two major limitations of existing GNNs in omics data analysis, i.e., limited-prediction (diagnosis) accuracy and limited-reproducible biomarker identification capacity across multiple datasets. The root of the challenges is the unique graph structure of biological signaling pathways, which consists of a large number of targets and intensive and complex signaling interactions among these targets. To resolve these two challenges, in this study, we presented a novel GNN model architecture, named PathFormer, which systematically integrate signaling network, priori knowledge and omics data to rank biomarkers and predict disease diagnosis. In the comparison results, PathFormer outperformed existing GNN models significantly in terms of highly accurate prediction capability ( 30% accuracy improvement in disease diagnosis compared with existing GNN models) and high reproducibility of biomarker ranking across different datasets. The improvement was confirmed using two independent Alzheimer's Disease (AD) and cancer transcriptomic datasets. The PathFormer model can be directly applied to other omics data analysis studies.
Universal Normalization Enhanced Graph Representation Learning for Gene Network Prediction
Sep 01, 2023



Abstract:Effective gene network representation learning is of great importance in bioinformatics to predict/understand the relation of gene profiles and disease phenotypes. Though graph neural networks (GNNs) have been the dominant architecture for analyzing various graph-structured data like social networks, their predicting on gene networks often exhibits subpar performance. In this paper, we formally investigate the gene network representation learning problem and characterize a notion of \textit{universal graph normalization}, where graph normalization can be applied in an universal manner to maximize the expressive power of GNNs while maintaining the stability. We propose a novel UNGNN (Universal Normalized GNN) framework, which leverages universal graph normalization in both the message passing phase and readout layer to enhance the performance of a base GNN. UNGNN has a plug-and-play property and can be combined with any GNN backbone in practice. A comprehensive set of experiments on gene-network-based bioinformatical tasks demonstrates that our UNGNN model significantly outperforms popular GNN benchmarks and provides an overall performance improvement of 16 $\%$ on average compared to previous state-of-the-art (SOTA) baselines. Furthermore, we also evaluate our theoretical findings on other graph datasets where the universal graph normalization is solvable, and we observe that UNGNN consistently achieves the superior performance.
Self-explaining Neural Network with Plausible Explanations
Oct 09, 2021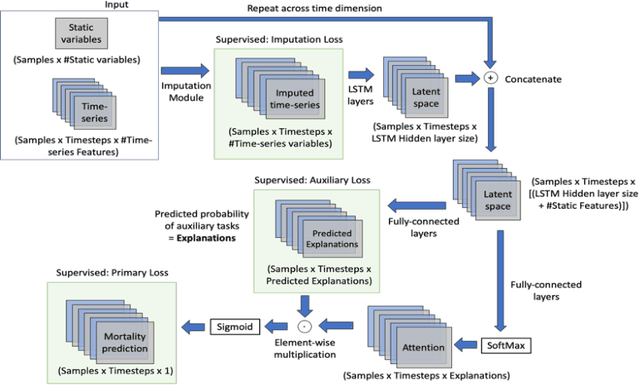
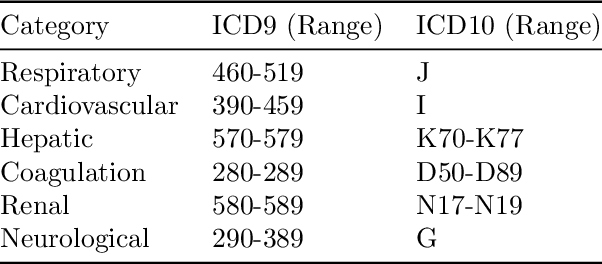
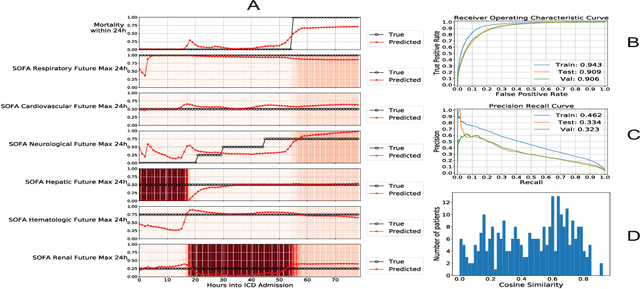
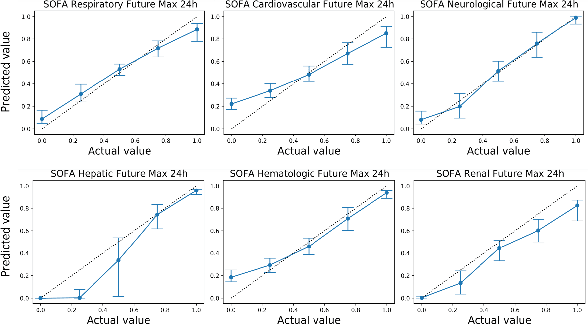
Abstract:Explaining the predictions of complex deep learning models, often referred to as black boxes, is critical in high-stakes domains like healthcare. However, post-hoc model explanations often are not understandable by clinicians and are difficult to integrate into clinical workflow. Further, while most explainable models use individual clinical variables as units of explanation, human understanding often rely on higher-level concepts or feature representations. In this paper, we propose a novel, self-explaining neural network for longitudinal in-hospital mortality prediction using domain-knowledge driven Sequential Organ Failure Assessment (SOFA) organ-specific scores as the atomic units of explanation. We also design a novel procedure to quantitatively validate the model explanations against gold standard discharge diagnosis information of patients. Our results provide interesting insights into how each of the SOFA organ scores contribute to mortality at different timesteps within longitudinal patient trajectory.
Synergistic Drug Combination Prediction by Integrating Multi-omics Data in Deep Learning Models
Nov 16, 2018
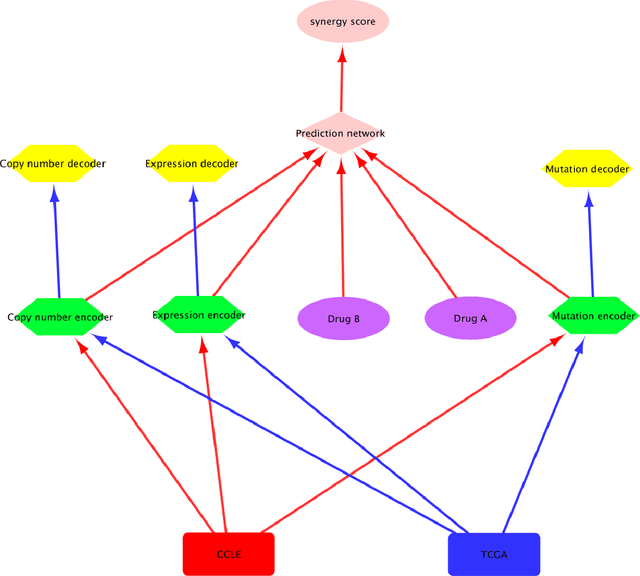
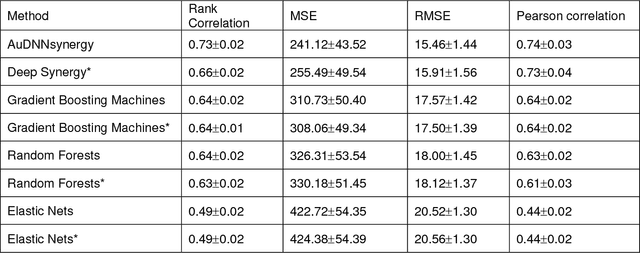
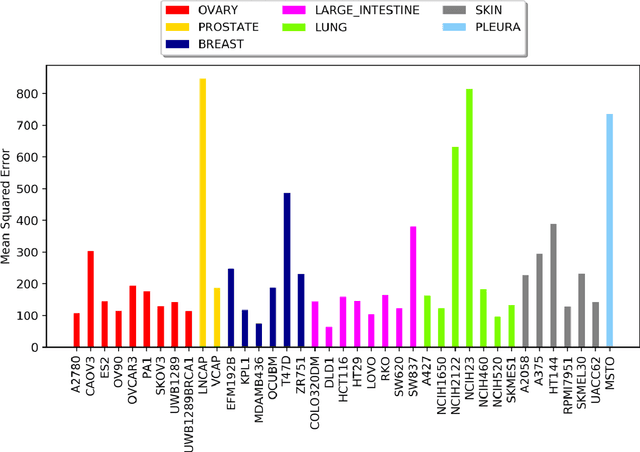
Abstract:Drug resistance is still a major challenge in cancer therapy. Drug combination is expected to overcome drug resistance. However, the number of possible drug combinations is enormous, and thus it is infeasible to experimentally screen all effective drug combinations considering the limited resources. Therefore, computational models to predict and prioritize effective drug combinations is important for combinatory therapy discovery in cancer. In this study, we proposed a novel deep learning model, AuDNNsynergy, to prediction drug combinations by integrating multi-omics data and chemical structure data. In specific, three autoencoders were trained using the gene expression, copy number and genetic mutation data of all tumor samples from The Cancer Genome Atlas. Then the physicochemical properties of drugs combined with the output of the three autoencoders, characterizing the individual cancer cell-lines, were used as the input of a deep neural network that predicts the synergy value of given pair-wise drug combinations against the specific cancer cell-lines. The comparison results showed the proposed AuDNNsynergy model outperforms four state-of-art approaches, namely DeepSynergy, Gradient Boosting Machines, Random Forests, and Elastic Nets. Moreover, we conducted the interpretation analysis of the deep learning model to investigate potential vital genetic predictors and the underlying mechanism of synergistic drug combinations on specific cancer cell-lines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge