Zehao Dong
SPGNN: Recognizing Salient Subgraph Patterns via Enhanced Graph Convolution and Pooling
Apr 29, 2024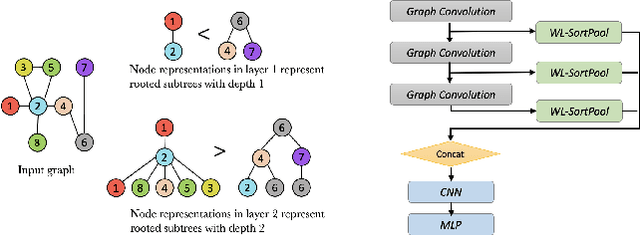
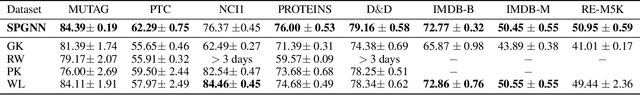
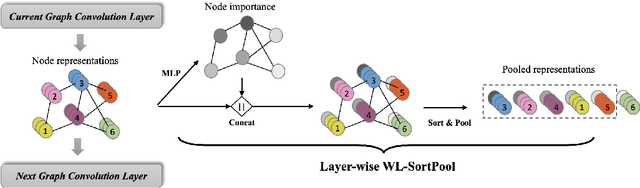
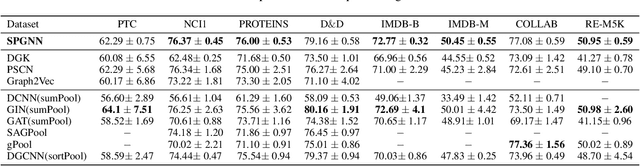
Abstract:Graph neural networks (GNNs) have revolutionized the field of machine learning on non-Euclidean data such as graphs and networks. GNNs effectively implement node representation learning through neighborhood aggregation and achieve impressive results in many graph-related tasks. However, most neighborhood aggregation approaches are summation-based, which can be problematic as they may not be sufficiently expressive to encode informative graph structures. Furthermore, though the graph pooling module is also of vital importance for graph learning, especially for the task of graph classification, research on graph down-sampling mechanisms is rather limited. To address the above challenges, we propose a concatenation-based graph convolution mechanism that injectively updates node representations to maximize the discriminative power in distinguishing non-isomorphic subgraphs. In addition, we design a novel graph pooling module, called WL-SortPool, to learn important subgraph patterns in a deep-learning manner. WL-SortPool layer-wise sorts node representations (i.e. continuous WL colors) to separately learn the relative importance of subtrees with different depths for the purpose of classification, thus better characterizing the complex graph topology and rich information encoded in the graph. We propose a novel Subgraph Pattern GNN (SPGNN) architecture that incorporates these enhancements. We test the proposed SPGNN architecture on many graph classification benchmarks. Experimental results show that our method can achieve highly competitive results with state-of-the-art graph kernels and other GNN approaches.
Large-Language-Model Empowered Dose Volume Histogram Prediction for Intensity Modulated Radiotherapy
Feb 11, 2024



Abstract:Treatment planning is currently a patient specific, time-consuming, and resource demanding task in radiotherapy. Dose-volume histogram (DVH) prediction plays a critical role in automating this process. The geometric relationship between DVHs in radiotherapy plans and organs-at-risk (OAR) and planning target volume (PTV) has been well established. This study explores the potential of deep learning models for predicting DVHs using images and subsequent human intervention facilitated by a large-language model (LLM) to enhance the planning quality. We propose a pipeline to convert unstructured images to a structured graph consisting of image-patch nodes and dose nodes. A novel Dose Graph Neural Network (DoseGNN) model is developed for predicting DVHs from the structured graph. The proposed DoseGNN is enhanced with the LLM to encode massive knowledge from prescriptions and interactive instructions from clinicians. In this study, we introduced an online human-AI collaboration (OHAC) system as a practical implementation of the concept proposed for the automation of intensity-modulated radiotherapy (IMRT) planning. In comparison to the widely-employed DL models used in radiotherapy, DoseGNN achieved mean square errors that were 80$\%$, 76$\%$ and 41.0$\%$ of those predicted by Swin U-Net Transformer, 3D U-Net CNN and vanilla MLP, respectively. Moreover, the LLM-empowered DoseGNN model facilitates seamless adjustment to treatment plans through interaction with clinicians using natural language.
Highly Accurate Disease Diagnosis and Highly Reproducible Biomarker Identification with PathFormer
Feb 11, 2024



Abstract:Biomarker identification is critical for precise disease diagnosis and understanding disease pathogenesis in omics data analysis, like using fold change and regression analysis. Graph neural networks (GNNs) have been the dominant deep learning model for analyzing graph-structured data. However, we found two major limitations of existing GNNs in omics data analysis, i.e., limited-prediction (diagnosis) accuracy and limited-reproducible biomarker identification capacity across multiple datasets. The root of the challenges is the unique graph structure of biological signaling pathways, which consists of a large number of targets and intensive and complex signaling interactions among these targets. To resolve these two challenges, in this study, we presented a novel GNN model architecture, named PathFormer, which systematically integrate signaling network, priori knowledge and omics data to rank biomarkers and predict disease diagnosis. In the comparison results, PathFormer outperformed existing GNN models significantly in terms of highly accurate prediction capability ( 30% accuracy improvement in disease diagnosis compared with existing GNN models) and high reproducibility of biomarker ranking across different datasets. The improvement was confirmed using two independent Alzheimer's Disease (AD) and cancer transcriptomic datasets. The PathFormer model can be directly applied to other omics data analysis studies.
DoseGNN: Improving the Performance of Deep Learning Models in Adaptive Dose-Volume Histogram Prediction through Graph Neural Networks
Feb 02, 2024Abstract:Dose-Volume Histogram (DVH) prediction is fundamental in radiation therapy that facilitate treatment planning, dose evaluation, plan comparison and etc. It helps to increase the ability to deliver precise and effective radiation treatments while managing potential toxicities to healthy tissues as needed to reduce the risk of complications. This paper extends recently disclosed research findings presented on AAPM (AAPM 65th Annual Meeting $\&$ Exhibition) and includes necessary technique details. The objective is to design efficient deep learning models for DVH prediction on general radiotherapy platform equipped with high performance CBCT system, where input CT images and target dose images to predict may have different origins, spacing and sizes. Deep learning models widely-adopted in DVH prediction task are evaluated on the novel radiotherapy platform, and graph neural networks (GNNs) are shown to be the ideal architecture to construct a plug-and-play framework to improve predictive performance of base deep learning models in the adaptive setting.
GNNHLS: Evaluating Graph Neural Network Inference via High-Level Synthesis
Sep 27, 2023Abstract:With the ever-growing popularity of Graph Neural Networks (GNNs), efficient GNN inference is gaining tremendous attention. Field-Programming Gate Arrays (FPGAs) are a promising execution platform due to their fine-grained parallelism, low-power consumption, reconfigurability, and concurrent execution. Even better, High-Level Synthesis (HLS) tools bridge the gap between the non-trivial FPGA development efforts and rapid emergence of new GNN models. In this paper, we propose GNNHLS, an open-source framework to comprehensively evaluate GNN inference acceleration on FPGAs via HLS, containing a software stack for data generation and baseline deployment, and FPGA implementations of 6 well-tuned GNN HLS kernels. We evaluate GNNHLS on 4 graph datasets with distinct topologies and scales. The results show that GNNHLS achieves up to 50.8x speedup and 423x energy reduction relative to the CPU baselines. Compared with the GPU baselines, GNNHLS achieves up to 5.16x speedup and 74.5x energy reduction.
Universal Normalization Enhanced Graph Representation Learning for Gene Network Prediction
Sep 01, 2023



Abstract:Effective gene network representation learning is of great importance in bioinformatics to predict/understand the relation of gene profiles and disease phenotypes. Though graph neural networks (GNNs) have been the dominant architecture for analyzing various graph-structured data like social networks, their predicting on gene networks often exhibits subpar performance. In this paper, we formally investigate the gene network representation learning problem and characterize a notion of \textit{universal graph normalization}, where graph normalization can be applied in an universal manner to maximize the expressive power of GNNs while maintaining the stability. We propose a novel UNGNN (Universal Normalized GNN) framework, which leverages universal graph normalization in both the message passing phase and readout layer to enhance the performance of a base GNN. UNGNN has a plug-and-play property and can be combined with any GNN backbone in practice. A comprehensive set of experiments on gene-network-based bioinformatical tasks demonstrates that our UNGNN model significantly outperforms popular GNN benchmarks and provides an overall performance improvement of 16 $\%$ on average compared to previous state-of-the-art (SOTA) baselines. Furthermore, we also evaluate our theoretical findings on other graph datasets where the universal graph normalization is solvable, and we observe that UNGNN consistently achieves the superior performance.
CktGNN: Circuit Graph Neural Network for Electronic Design Automation
Aug 31, 2023Abstract:The electronic design automation of analog circuits has been a longstanding challenge in the integrated circuit field due to the huge design space and complex design trade-offs among circuit specifications. In the past decades, intensive research efforts have mostly been paid to automate the transistor sizing with a given circuit topology. By recognizing the graph nature of circuits, this paper presents a Circuit Graph Neural Network (CktGNN) that simultaneously automates the circuit topology generation and device sizing based on the encoder-dependent optimization subroutines. Particularly, CktGNN encodes circuit graphs using a two-level GNN framework (of nested GNN) where circuits are represented as combinations of subgraphs in a known subgraph basis. In this way, it significantly improves design efficiency by reducing the number of subgraphs to perform message passing. Nonetheless, another critical roadblock to advancing learning-assisted circuit design automation is a lack of public benchmarks to perform canonical assessment and reproducible research. To tackle the challenge, we introduce Open Circuit Benchmark (OCB), an open-sourced dataset that contains $10$K distinct operational amplifiers with carefully-extracted circuit specifications. OCB is also equipped with communicative circuit generation and evaluation capabilities such that it can help to generalize CktGNN to design various analog circuits by producing corresponding datasets. Experiments on OCB show the extraordinary advantages of CktGNN through representation-based optimization frameworks over other recent powerful GNN baselines and human experts' manual designs. Our work paves the way toward a learning-based open-sourced design automation for analog circuits. Our source code is available at \url{https://github.com/zehao-dong/CktGNN}.
Time Associated Meta Learning for Clinical Prediction
Mar 05, 2023Abstract:Rich Electronic Health Records (EHR), have created opportunities to improve clinical processes using machine learning methods. Prediction of the same patient events at different time horizons can have very different applications and interpretations; however, limited number of events in each potential time window hurts the effectiveness of conventional machine learning algorithms. We propose a novel time associated meta learning (TAML) method to make effective predictions at multiple future time points. We view time-associated disease prediction as classification tasks at multiple time points. Such closely-related classification tasks are an excellent candidate for model-based meta learning. To address the sparsity problem after task splitting, TAML employs a temporal information sharing strategy to augment the number of positive samples and include the prediction of related phenotypes or events in the meta-training phase. We demonstrate the effectiveness of TAML on multiple clinical datasets, where it consistently outperforms a range of strong baselines. We also develop a MetaEHR package for implementing both time-associated and time-independent few-shot prediction on EHR data.
Interpreting mechanism of Synergism of drug combinations using attention based hierarchical graph pooling
Sep 19, 2022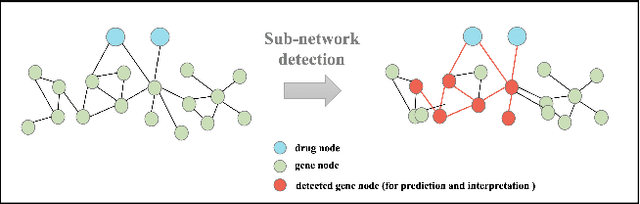
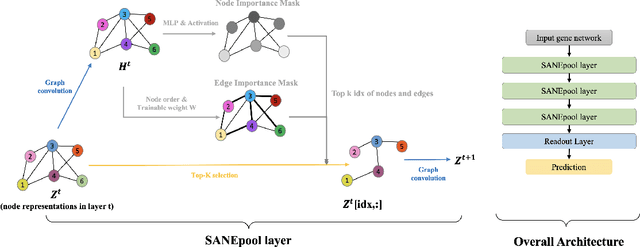
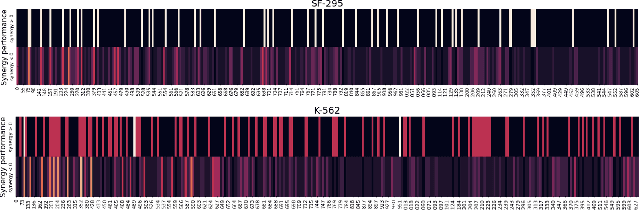
Abstract:The synergistic drug combinations provide huge potentials to enhance therapeutic efficacy and to reduce adverse reactions. However, effective and synergistic drug combination prediction remains an open question because of the unknown causal disease signaling pathways. Though various deep learning (AI) models have been proposed to quantitatively predict the synergism of drug combinations. The major limitation of existing deep learning methods is that they are inherently not interpretable, which makes the conclusion of AI models un-transparent to human experts, henceforth limiting the robustness of the model conclusion and the implementation ability of these models in the real-world human-AI healthcare. In this paper, we develop an interpretable graph neural network (GNN) that reveals the underlying essential therapeutic targets and mechanism of the synergy (MoS) by mining the sub-molecular network of great importance. The key point of the interpretable GNN prediction model is a novel graph pooling layer, Self-Attention based Node and Edge pool (henceforth SANEpool), that can compute the attention score (importance) of nodes and edges based on the node features and the graph topology. As such, the proposed GNN model provides a systematic way to predict and interpret the drug combination synergism based on the detected crucial sub-molecular network. We evaluate SANEpool on molecular networks formulated by genes from 46 core cancer signaling pathways and drug combinations from NCI ALMANAC drug combination screening data. The experimental results indicate that 1) SANEpool can achieve the current state-of-art performance among other popular graph neural networks; and 2) the sub-molecular network detected by SANEpool are self-explainable and salient for identifying synergistic drug combinations.
PACE: A Parallelizable Computation Encoder for Directed Acyclic Graphs
Mar 19, 2022
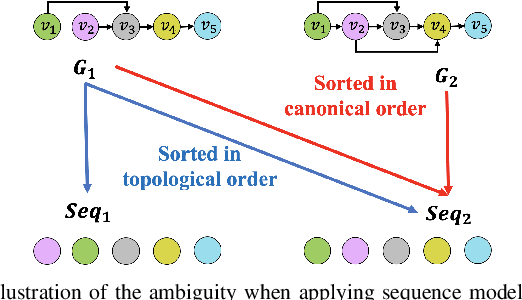
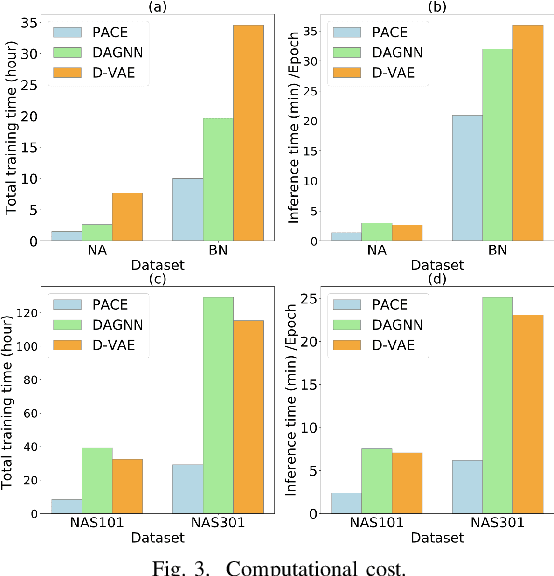

Abstract:Optimization of directed acyclic graph (DAG) structures has many applications, such as neural architecture search (NAS) and probabilistic graphical model learning. Encoding DAGs into real vectors is a dominant component in most neural-network-based DAG optimization frameworks. Currently, most DAG encoders use an asynchronous message passing scheme which sequentially processes nodes according to the dependency between nodes in a DAG. That is, a node must not be processed until all its predecessors are processed. As a result, they are inherently not parallelizable. In this work, we propose a Parallelizable Attention-based Computation structure Encoder (PACE) that processes nodes simultaneously and encodes DAGs in parallel. We demonstrate the superiority of PACE through encoder-dependent optimization subroutines that search the optimal DAG structure based on the learned DAG embeddings. Experiments show that PACE not only improves the effectiveness over previous sequential DAG encoders with a significantly boosted training and inference speed, but also generates smooth latent (DAG encoding) spaces that are beneficial to downstream optimization subroutines. Our source code is available at \url{https://github.com/zehaodong/PACE}
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge