Interpreting mechanism of Synergism of drug combinations using attention based hierarchical graph pooling
Paper and Code
Sep 19, 2022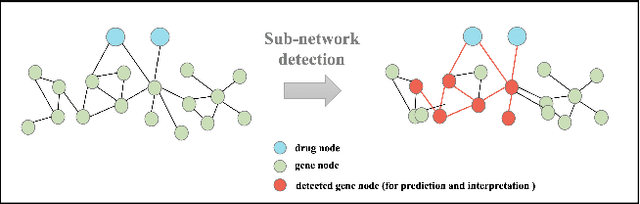
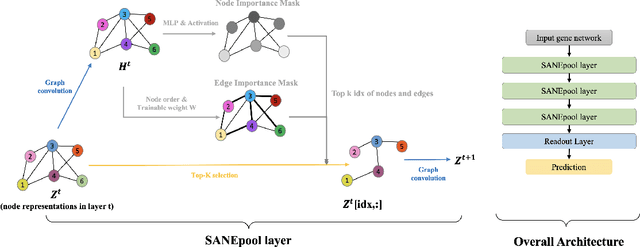
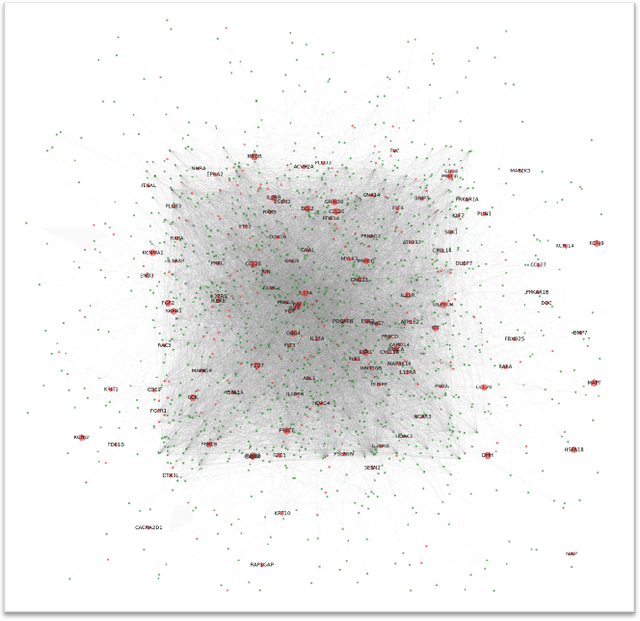
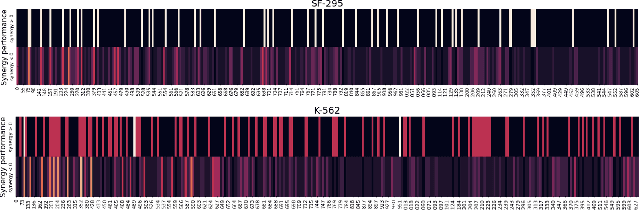
The synergistic drug combinations provide huge potentials to enhance therapeutic efficacy and to reduce adverse reactions. However, effective and synergistic drug combination prediction remains an open question because of the unknown causal disease signaling pathways. Though various deep learning (AI) models have been proposed to quantitatively predict the synergism of drug combinations. The major limitation of existing deep learning methods is that they are inherently not interpretable, which makes the conclusion of AI models un-transparent to human experts, henceforth limiting the robustness of the model conclusion and the implementation ability of these models in the real-world human-AI healthcare. In this paper, we develop an interpretable graph neural network (GNN) that reveals the underlying essential therapeutic targets and mechanism of the synergy (MoS) by mining the sub-molecular network of great importance. The key point of the interpretable GNN prediction model is a novel graph pooling layer, Self-Attention based Node and Edge pool (henceforth SANEpool), that can compute the attention score (importance) of nodes and edges based on the node features and the graph topology. As such, the proposed GNN model provides a systematic way to predict and interpret the drug combination synergism based on the detected crucial sub-molecular network. We evaluate SANEpool on molecular networks formulated by genes from 46 core cancer signaling pathways and drug combinations from NCI ALMANAC drug combination screening data. The experimental results indicate that 1) SANEpool can achieve the current state-of-art performance among other popular graph neural networks; and 2) the sub-molecular network detected by SANEpool are self-explainable and salient for identifying synergistic drug combinations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge