Philip Payne
OmniCellTOSG: The First Cell Text-Omic Signaling Graphs Dataset for Joint LLM and GNN Modeling
Apr 02, 2025Abstract:Complex cell signaling systems -- governed by varying protein abundances and interactions -- generate diverse cell types across organs. These systems evolve under influences such as age, sex, diet, environmental exposures, and diseases, making them challenging to decode given the involvement of tens of thousands of genes and proteins. Recently, hundreds of millions of single-cell omics data have provided a robust foundation for understanding these signaling networks within various cell subpopulations and conditions. Inspired by the success of large foundation models (for example, large language models and large vision models) pre-trained on massive datasets, we introduce OmniCellTOSG, the first dataset of cell text-omic signaling graphs (TOSGs). Each TOSG represents the signaling network of an individual or meta-cell and is labeled with information such as organ, disease, sex, age, and cell subtype. OmniCellTOSG offers two key contributions. First, it introduces a novel graph model that integrates human-readable annotations -- such as biological functions, cellular locations, signaling pathways, related diseases, and drugs -- with quantitative gene and protein abundance data, enabling graph reasoning to decode cell signaling. This approach calls for new joint models combining large language models and graph neural networks. Second, the dataset is built from single-cell RNA sequencing data of approximately 120 million cells from diverse tissues and conditions (healthy and diseased) and is fully compatible with PyTorch. This facilitates the development of innovative cell signaling models that could transform research in life sciences, healthcare, and precision medicine. The OmniCellTOSG dataset is continuously expanding and will be updated regularly. The dataset and code are available at https://github.com/FuhaiLiAiLab/OmniCellTOSG.
KoGNER: A Novel Framework for Knowledge Graph Distillation on Biomedical Named Entity Recognition
Mar 19, 2025Abstract:Named Entity Recognition (NER) is a fundamental task in Natural Language Processing (NLP) that plays a crucial role in information extraction, question answering, and knowledge-based systems. Traditional deep learning-based NER models often struggle with domain-specific generalization and suffer from data sparsity issues. In this work, we introduce Knowledge Graph distilled for Named Entity Recognition (KoGNER), a novel approach that integrates Knowledge Graph (KG) distillation into NER models to enhance entity recognition performance. Our framework leverages structured knowledge representations from KGs to enrich contextual embeddings, thereby improving entity classification and reducing ambiguity in entity detection. KoGNER employs a two-step process: (1) Knowledge Distillation, where external knowledge sources are distilled into a lightweight representation for seamless integration with NER models, and (2) Entity-Aware Augmentation, which integrates contextual embeddings that have been enriched with knowledge graph information directly into GNN, thereby improving the model's ability to understand and represent entity relationships. Experimental results on benchmark datasets demonstrate that KoGNER achieves state-of-the-art performance, outperforming finetuned NER models and LLMs by a significant margin. These findings suggest that leveraging knowledge graphs as auxiliary information can significantly improve NER accuracy, making KoGNER a promising direction for future research in knowledge-aware NLP.
Analysing heterogeneity in Alzheimer Disease using multimodal normative modelling on ATN biomarkers
Apr 04, 2024Abstract:Alzheimer Disease (AD) is a multi-faceted disorder, with each modality providing unique and complementary info about AD. In this study, we used a deep-learning based multimodal normative model to assess the heterogeneity in regional brain patterns for ATN (amyloid-tau-neurodegeneration) biomarkers. We selected discovery (n = 665) and replication (n = 430) cohorts with simultaneous availability of ATN biomarkers: Florbetapir amyloid, Flortaucipir tau and T1-weighted MRI (magnetic resonance imaging) imaging. A multimodal variational autoencoder (conditioned on age and sex) was used as a normative model to learn the multimodal regional brain patterns of a cognitively unimpaired (CU) control group. The trained model was applied on individuals on the ADS (AD Spectrum) to estimate their deviations (Z-scores) from the normative distribution, resulting in a Z-score regional deviation map per ADS individual per modality. ADS individuals with moderate or severe dementia showed higher proportion of regional outliers for each modality as well as more dissimilarity in modality-specific regional outlier patterns compared to ADS individuals with early or mild dementia. DSI was associated with the progressive stages of dementia, (ii) showed significant associations with neuropsychological composite scores and (iii) related to the longitudinal risk of CDR progression. Findings were reproducible in both discovery and replication cohorts. Our is the first study to examine the heterogeneity in AD through the lens of multiple neuroimaging modalities (ATN), based on distinct or overlapping patterns of regional outlier deviations. Regional MRI and tau outliers were more heterogenous than regional amyloid outliers. DSI has the potential to be an individual patient metric of neurodegeneration that can help in clinical decision making and monitoring patient response for anti-amyloid treatments.
Multimodal hierarchical multi-task deep learning framework for jointly predicting and explaining Alzheimer disease progression
Apr 04, 2024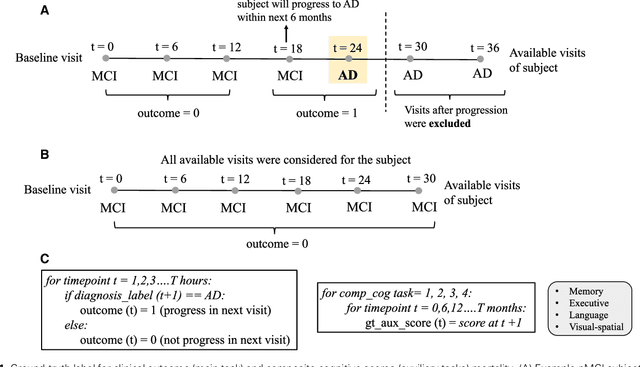
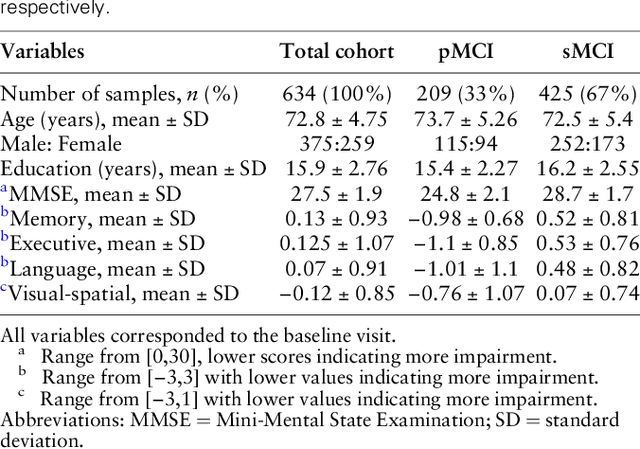
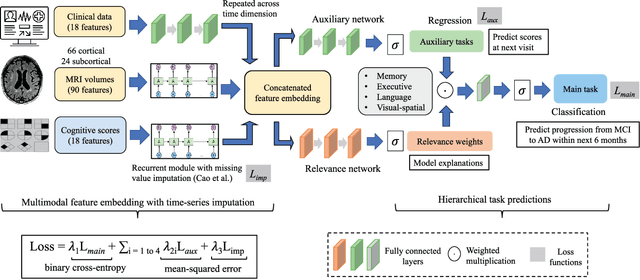
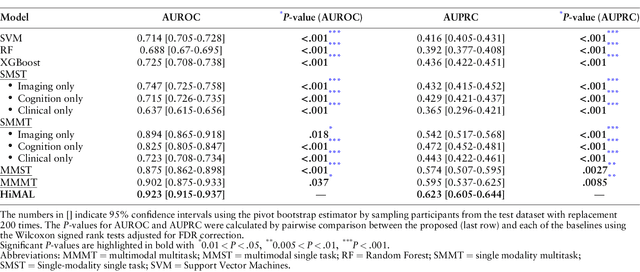
Abstract:Early identification of Mild Cognitive Impairment (MCI) subjects who will eventually progress to Alzheimer Disease (AD) is challenging. Existing deep learning models are mostly single-modality single-task models predicting risk of disease progression at a fixed timepoint. We proposed a multimodal hierarchical multi-task learning approach which can monitor the risk of disease progression at each timepoint of the visit trajectory. Longitudinal visit data from multiple modalities (MRI, cognition, and clinical data) were collected from MCI individuals of the Alzheimer Disease Neuroimaging Initiative (ADNI) dataset. Our hierarchical model predicted at every timepoint a set of neuropsychological composite cognitive function scores as auxiliary tasks and used the forecasted scores at every timepoint to predict the future risk of disease. Relevance weights for each composite function provided explanations about potential factors for disease progression. Our proposed model performed better than state-of-the-art baselines in predicting AD progression risk and the composite scores. Ablation study on the number of modalities demonstrated that imaging and cognition data contributed most towards the outcome. Model explanations at each timepoint can inform clinicians 6 months in advance the potential cognitive function decline that can lead to progression to AD in future. Our model monitored their risk of AD progression every 6 months throughout the visit trajectory of individuals. The hierarchical learning of auxiliary tasks allowed better optimization and allowed longitudinal explanations for the outcome. Our framework is flexible with the number of input modalities and the selection of auxiliary tasks and hence can be generalized to other clinical problems too.
Improving Normative Modeling for Multi-modal Neuroimaging Data using mixture-of-product-of-experts variational autoencoders
Dec 02, 2023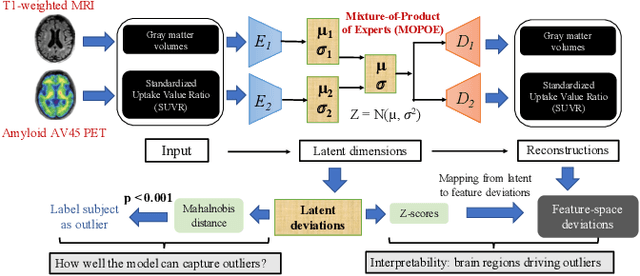
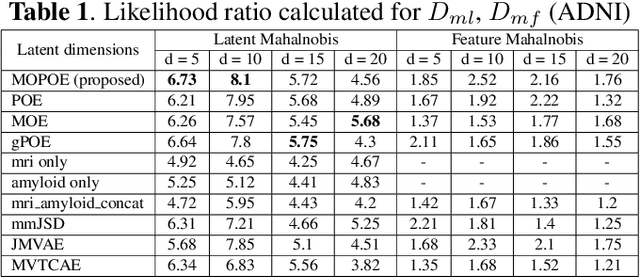
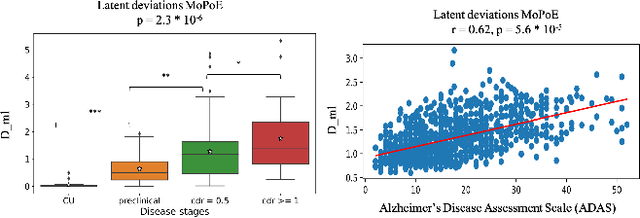
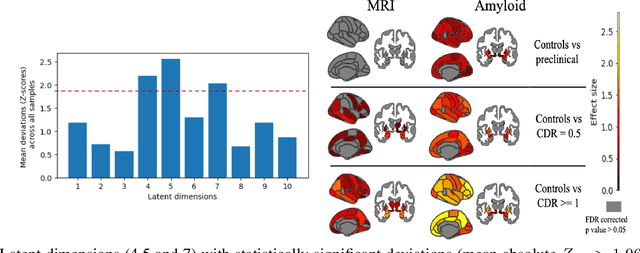
Abstract:Normative models in neuroimaging learn the brain patterns of healthy population distribution and estimate how disease subjects like Alzheimer's Disease (AD) deviate from the norm. Existing variational autoencoder (VAE)-based normative models using multimodal neuroimaging data aggregate information from multiple modalities by estimating product or averaging of unimodal latent posteriors. This can often lead to uninformative joint latent distributions which affects the estimation of subject-level deviations. In this work, we addressed the prior limitations by adopting the Mixture-of-Product-of-Experts (MoPoE) technique which allows better modelling of the joint latent posterior. Our model labelled subjects as outliers by calculating deviations from the multimodal latent space. Further, we identified which latent dimensions and brain regions were associated with abnormal deviations due to AD pathology.
Assisting Clinical Decisions for Scarcely Available Treatment via Disentangled Latent Representation
Jul 06, 2023



Abstract:Extracorporeal membrane oxygenation (ECMO) is an essential life-supporting modality for COVID-19 patients who are refractory to conventional therapies. However, the proper treatment decision has been the subject of significant debate and it remains controversial about who benefits from this scarcely available and technically complex treatment option. To support clinical decisions, it is a critical need to predict the treatment need and the potential treatment and no-treatment responses. Targeting this clinical challenge, we propose Treatment Variational AutoEncoder (TVAE), a novel approach for individualized treatment analysis. TVAE is specifically designed to address the modeling challenges like ECMO with strong treatment selection bias and scarce treatment cases. TVAE conceptualizes the treatment decision as a multi-scale problem. We model a patient's potential treatment assignment and the factual and counterfactual outcomes as part of their intrinsic characteristics that can be represented by a deep latent variable model. The factual and counterfactual prediction errors are alleviated via a reconstruction regularization scheme together with semi-supervision, and the selection bias and the scarcity of treatment cases are mitigated by the disentangled and distribution-matched latent space and the label-balancing generative strategy. We evaluate TVAE on two real-world COVID-19 datasets: an international dataset collected from 1651 hospitals across 63 countries, and a institutional dataset collected from 15 hospitals. The results show that TVAE outperforms state-of-the-art treatment effect models in predicting both the propensity scores and factual outcomes on heterogeneous COVID-19 datasets. Additional experiments also show TVAE outperforms the best existing models in individual treatment effect estimation on the synthesized IHDP benchmark dataset.
Interpreting mechanism of Synergism of drug combinations using attention based hierarchical graph pooling
Sep 19, 2022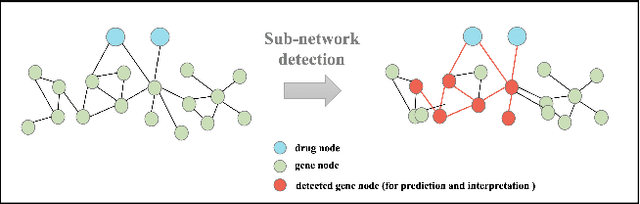
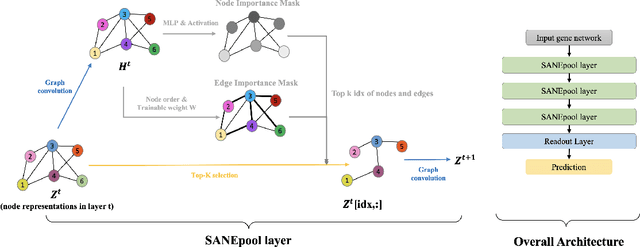
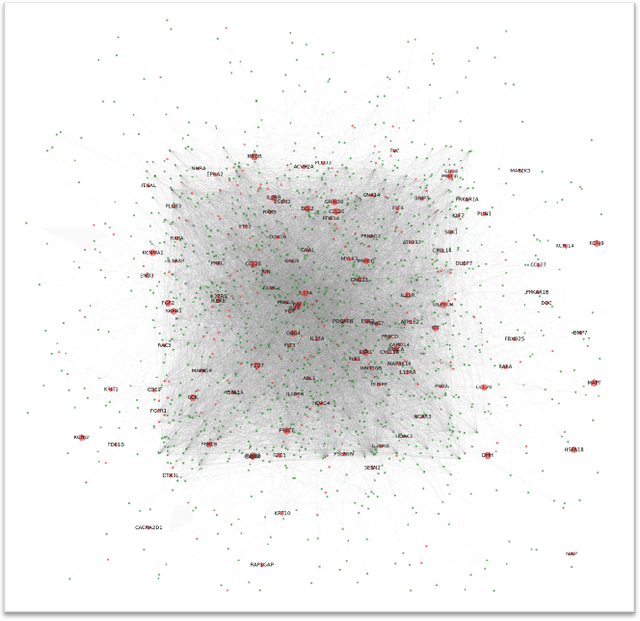
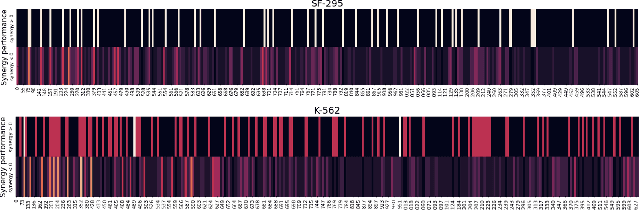
Abstract:The synergistic drug combinations provide huge potentials to enhance therapeutic efficacy and to reduce adverse reactions. However, effective and synergistic drug combination prediction remains an open question because of the unknown causal disease signaling pathways. Though various deep learning (AI) models have been proposed to quantitatively predict the synergism of drug combinations. The major limitation of existing deep learning methods is that they are inherently not interpretable, which makes the conclusion of AI models un-transparent to human experts, henceforth limiting the robustness of the model conclusion and the implementation ability of these models in the real-world human-AI healthcare. In this paper, we develop an interpretable graph neural network (GNN) that reveals the underlying essential therapeutic targets and mechanism of the synergy (MoS) by mining the sub-molecular network of great importance. The key point of the interpretable GNN prediction model is a novel graph pooling layer, Self-Attention based Node and Edge pool (henceforth SANEpool), that can compute the attention score (importance) of nodes and edges based on the node features and the graph topology. As such, the proposed GNN model provides a systematic way to predict and interpret the drug combination synergism based on the detected crucial sub-molecular network. We evaluate SANEpool on molecular networks formulated by genes from 46 core cancer signaling pathways and drug combinations from NCI ALMANAC drug combination screening data. The experimental results indicate that 1) SANEpool can achieve the current state-of-art performance among other popular graph neural networks; and 2) the sub-molecular network detected by SANEpool are self-explainable and salient for identifying synergistic drug combinations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge