Tanya Berger-Wolf
A continental-scale dataset of ground beetles with high-resolution images and validated morphological trait measurements
Jan 14, 2026Abstract:Despite the ecological significance of invertebrates, global trait databases remain heavily biased toward vertebrates and plants, limiting comprehensive ecological analyses of high-diversity groups like ground beetles. Ground beetles (Coleoptera: Carabidae) serve as critical bioindicators of ecosystem health, providing valuable insights into biodiversity shifts driven by environmental changes. While the National Ecological Observatory Network (NEON) maintains an extensive collection of carabid specimens from across the United States, these primarily exist as physical collections, restricting widespread research access and large-scale analysis. To address these gaps, we present a multimodal dataset digitizing over 13,200 NEON carabids from 30 sites spanning the continental US and Hawaii through high-resolution imaging, enabling broader access and computational analysis. The dataset includes digitally measured elytra length and width of each specimen, establishing a foundation for automated trait extraction using AI. Validated against manual measurements, our digital trait extraction achieves sub-millimeter precision, ensuring reliability for ecological and computational studies. By addressing invertebrate under-representation in trait databases, this work supports AI-driven tools for automated species identification and trait-based research, fostering advancements in biodiversity monitoring and conservation.
kabr-tools: Automated Framework for Multi-Species Behavioral Monitoring
Oct 02, 2025Abstract:A comprehensive understanding of animal behavior ecology depends on scalable approaches to quantify and interpret complex, multidimensional behavioral patterns. Traditional field observations are often limited in scope, time-consuming, and labor-intensive, hindering the assessment of behavioral responses across landscapes. To address this, we present kabr-tools (Kenyan Animal Behavior Recognition Tools), an open-source package for automated multi-species behavioral monitoring. This framework integrates drone-based video with machine learning systems to extract behavioral, social, and spatial metrics from wildlife footage. Our pipeline leverages object detection, tracking, and behavioral classification systems to generate key metrics, including time budgets, behavioral transitions, social interactions, habitat associations, and group composition dynamics. Compared to ground-based methods, drone-based observations significantly improved behavioral granularity, reducing visibility loss by 15% and capturing more transitions with higher accuracy and continuity. We validate kabr-tools through three case studies, analyzing 969 behavioral sequences, surpassing the capacity of traditional methods for data capture and annotation. We found that, like Plains zebras, vigilance in Grevy's zebras decreases with herd size, but, unlike Plains zebras, habitat has a negligible impact. Plains and Grevy's zebras exhibit strong behavioral inertia, with rare transitions to alert behaviors and observed spatial segregation between Grevy's zebras, Plains zebras, and giraffes in mixed-species herds. By enabling automated behavioral monitoring at scale, kabr-tools offers a powerful tool for ecosystem-wide studies, advancing conservation, biodiversity research, and ecological monitoring.
BioCLIP 2: Emergent Properties from Scaling Hierarchical Contrastive Learning
May 29, 2025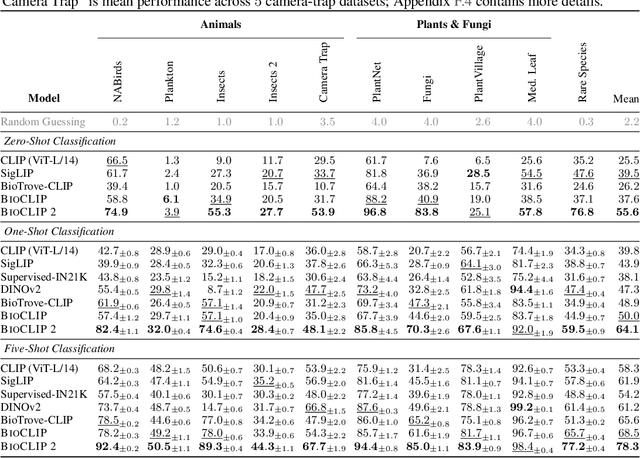
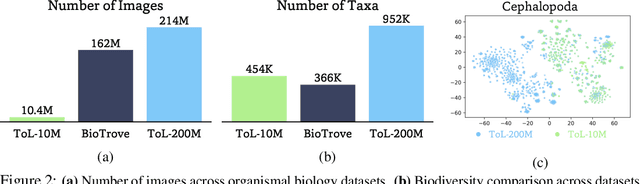
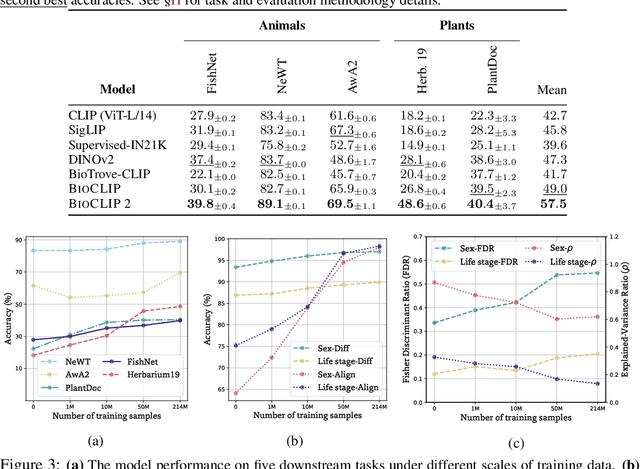
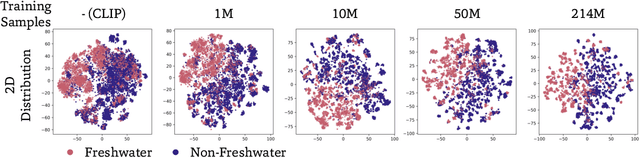
Abstract:Foundation models trained at scale exhibit remarkable emergent behaviors, learning new capabilities beyond their initial training objectives. We find such emergent behaviors in biological vision models via large-scale contrastive vision-language training. To achieve this, we first curate TreeOfLife-200M, comprising 214 million images of living organisms, the largest and most diverse biological organism image dataset to date. We then train BioCLIP 2 on TreeOfLife-200M to distinguish different species. Despite the narrow training objective, BioCLIP 2 yields extraordinary accuracy when applied to various biological visual tasks such as habitat classification and trait prediction. We identify emergent properties in the learned embedding space of BioCLIP 2. At the inter-species level, the embedding distribution of different species aligns closely with functional and ecological meanings (e.g., beak sizes and habitats). At the intra-species level, instead of being diminished, the intra-species variations (e.g., life stages and sexes) are preserved and better separated in subspaces orthogonal to inter-species distinctions. We provide formal proof and analyses to explain why hierarchical supervision and contrastive objectives encourage these emergent properties. Crucially, our results reveal that these properties become increasingly significant with larger-scale training data, leading to a biologically meaningful embedding space.
Optimizing Image Capture for Computer Vision-Powered Taxonomic Identification and Trait Recognition of Biodiversity Specimens
May 22, 2025Abstract:Biological collections house millions of specimens documenting Earth's biodiversity, with digital images increasingly available through open-access platforms. Most imaging protocols were developed for human visual interpretation without considering computational analysis requirements. This paper aims to bridge the gap between current imaging practices and the potential for automated analysis by presenting key considerations for creating biological specimen images optimized for computer vision applications. We provide conceptual computer vision topics for context, addressing fundamental concerns including model generalization, data leakage, and comprehensive metadata documentation, and outline practical guidance on specimen imagine, and data storage. These recommendations were synthesized through interdisciplinary collaboration between taxonomists, collection managers, ecologists, and computer scientists. Through this synthesis, we have identified ten interconnected considerations that form a framework for successfully integrating biological specimen images into computer vision pipelines. The key elements include: (1) comprehensive metadata documentation, (2) standardized specimen positioning, (3) consistent size and color calibration, (4) protocols for handling multiple specimens in one image, (5) uniform background selection, (6) controlled lighting, (7) appropriate resolution and magnification, (8) optimal file formats, (9) robust data archiving strategies, and (10) accessible data sharing practices. By implementing these recommendations, collection managers, taxonomists, and biodiversity informaticians can generate images that support automated trait extraction, species identification, and novel ecological and evolutionary analyses at unprecedented scales. Successful implementation lies in thorough documentation of methodological choices.
Building Machine Learning Challenges for Anomaly Detection in Science
Mar 03, 2025



Abstract:Scientific discoveries are often made by finding a pattern or object that was not predicted by the known rules of science. Oftentimes, these anomalous events or objects that do not conform to the norms are an indication that the rules of science governing the data are incomplete, and something new needs to be present to explain these unexpected outliers. The challenge of finding anomalies can be confounding since it requires codifying a complete knowledge of the known scientific behaviors and then projecting these known behaviors on the data to look for deviations. When utilizing machine learning, this presents a particular challenge since we require that the model not only understands scientific data perfectly but also recognizes when the data is inconsistent and out of the scope of its trained behavior. In this paper, we present three datasets aimed at developing machine learning-based anomaly detection for disparate scientific domains covering astrophysics, genomics, and polar science. We present the different datasets along with a scheme to make machine learning challenges around the three datasets findable, accessible, interoperable, and reusable (FAIR). Furthermore, we present an approach that generalizes to future machine learning challenges, enabling the possibility of large, more compute-intensive challenges that can ultimately lead to scientific discovery.
Sparse Autoencoders for Scientifically Rigorous Interpretation of Vision Models
Feb 10, 2025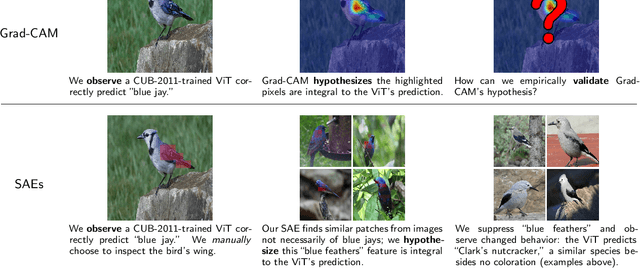

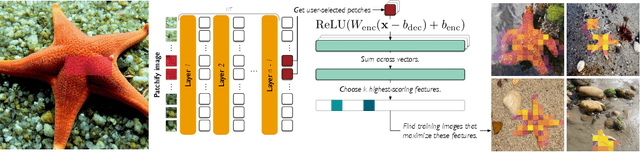
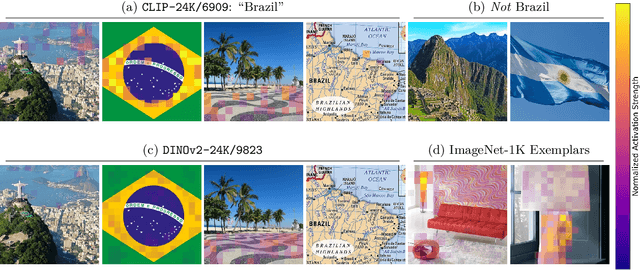
Abstract:To truly understand vision models, we must not only interpret their learned features but also validate these interpretations through controlled experiments. Current approaches either provide interpretable features without the ability to test their causal influence, or enable model editing without interpretable controls. We present a unified framework using sparse autoencoders (SAEs) that bridges this gap, allowing us to discover human-interpretable visual features and precisely manipulate them to test hypotheses about model behavior. By applying our method to state-of-the-art vision models, we reveal key differences in the semantic abstractions learned by models with different pre-training objectives. We then demonstrate the practical usage of our framework through controlled interventions across multiple vision tasks. We show that SAEs can reliably identify and manipulate interpretable visual features without model re-training, providing a powerful tool for understanding and controlling vision model behavior. We provide code, demos and models on our project website: https://osu-nlp-group.github.io/SAE-V.
Finer-CAM: Spotting the Difference Reveals Finer Details for Visual Explanation
Jan 20, 2025



Abstract:Class activation map (CAM) has been widely used to highlight image regions that contribute to class predictions. Despite its simplicity and computational efficiency, CAM often struggles to identify discriminative regions that distinguish visually similar fine-grained classes. Prior efforts address this limitation by introducing more sophisticated explanation processes, but at the cost of extra complexity. In this paper, we propose Finer-CAM, a method that retains CAM's efficiency while achieving precise localization of discriminative regions. Our key insight is that the deficiency of CAM lies not in "how" it explains, but in "what" it explains}. Specifically, previous methods attempt to identify all cues contributing to the target class's logit value, which inadvertently also activates regions predictive of visually similar classes. By explicitly comparing the target class with similar classes and spotting their differences, Finer-CAM suppresses features shared with other classes and emphasizes the unique, discriminative details of the target class. Finer-CAM is easy to implement, compatible with various CAM methods, and can be extended to multi-modal models for accurate localization of specific concepts. Additionally, Finer-CAM allows adjustable comparison strength, enabling users to selectively highlight coarse object contours or fine discriminative details. Quantitatively, we show that masking out the top 5% of activated pixels by Finer-CAM results in a larger relative confidence drop compared to baselines. The source code and demo are available at https://github.com/Imageomics/Finer-CAM.
Prompt-CAM: A Simpler Interpretable Transformer for Fine-Grained Analysis
Jan 16, 2025



Abstract:We present a simple usage of pre-trained Vision Transformers (ViTs) for fine-grained analysis, aiming to identify and localize the traits that distinguish visually similar categories, such as different bird species or dog breeds. Pre-trained ViTs such as DINO have shown remarkable capabilities to extract localized, informative features. However, using saliency maps like Grad-CAM can hardly point out the traits: they often locate the whole object by a blurred, coarse heatmap, not traits. We propose a novel approach Prompt Class Attention Map (Prompt-CAM) to the rescue. Prompt-CAM learns class-specific prompts to a pre-trained ViT and uses the corresponding outputs for classification. To classify an image correctly, the true-class prompt must attend to the unique image patches not seen in other classes' images, i.e., traits. As such, the true class's multi-head attention maps reveal traits and their locations. Implementation-wise, Prompt-CAM is almost a free lunch by simply modifying the prediction head of Visual Prompt Tuning (VPT). This makes Prompt-CAM fairly easy to train and apply, sharply contrasting other interpretable methods that design specific models and training processes. It is even simpler than the recently published INterpretable TRansformer (INTR), whose encoder-decoder architecture prevents it from leveraging pre-trained ViTs. Extensive empirical studies on a dozen datasets from various domains (e.g., birds, fishes, insects, fungi, flowers, food, and cars) validate Prompt-CAM superior interpretation capability.
Static Segmentation by Tracking: A Frustratingly Label-Efficient Approach to Fine-Grained Segmentation
Jan 12, 2025



Abstract:We study image segmentation in the biological domain, particularly trait and part segmentation from specimen images (e.g., butterfly wing stripes or beetle body parts). This is a crucial, fine-grained task that aids in understanding the biology of organisms. The conventional approach involves hand-labeling masks, often for hundreds of images per species, and training a segmentation model to generalize these labels to other images, which can be exceedingly laborious. We present a label-efficient method named Static Segmentation by Tracking (SST). SST is built upon the insight: while specimens of the same species have inherent variations, the traits and parts we aim to segment show up consistently. This motivates us to concatenate specimen images into a ``pseudo-video'' and reframe trait and part segmentation as a tracking problem. Concretely, SST generates masks for unlabeled images by propagating annotated or predicted masks from the ``pseudo-preceding'' images. Powered by Segment Anything Model 2 (SAM~2) initially developed for video segmentation, we show that SST can achieve high-quality trait and part segmentation with merely one labeled image per species -- a breakthrough for analyzing specimen images. We further develop a cycle-consistent loss to fine-tune the model, again using one labeled image. Additionally, we highlight the broader potential of SST, including one-shot instance segmentation on images taken in the wild and trait-based image retrieval.
Fine-Tuning is Fine, if Calibrated
Sep 24, 2024



Abstract:Fine-tuning is arguably the most straightforward way to tailor a pre-trained model (e.g., a foundation model) to downstream applications, but it also comes with the risk of losing valuable knowledge the model had learned in pre-training. For example, fine-tuning a pre-trained classifier capable of recognizing a large number of classes to master a subset of classes at hand is shown to drastically degrade the model's accuracy in the other classes it had previously learned. As such, it is hard to further use the fine-tuned model when it encounters classes beyond the fine-tuning data. In this paper, we systematically dissect the issue, aiming to answer the fundamental question, ''What has been damaged in the fine-tuned model?'' To our surprise, we find that the fine-tuned model neither forgets the relationship among the other classes nor degrades the features to recognize these classes. Instead, the fine-tuned model often produces more discriminative features for these other classes, even if they were missing during fine-tuning! {What really hurts the accuracy is the discrepant logit scales between the fine-tuning classes and the other classes}, implying that a simple post-processing calibration would bring back the pre-trained model's capability and at the same time unveil the feature improvement over all classes. We conduct an extensive empirical study to demonstrate the robustness of our findings and provide preliminary explanations underlying them, suggesting new directions for future theoretical analysis. Our code is available at https://github.com/OSU-MLB/Fine-Tuning-Is-Fine-If-Calibrated.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge