Mridul Khurana
A continental-scale dataset of ground beetles with high-resolution images and validated morphological trait measurements
Jan 14, 2026Abstract:Despite the ecological significance of invertebrates, global trait databases remain heavily biased toward vertebrates and plants, limiting comprehensive ecological analyses of high-diversity groups like ground beetles. Ground beetles (Coleoptera: Carabidae) serve as critical bioindicators of ecosystem health, providing valuable insights into biodiversity shifts driven by environmental changes. While the National Ecological Observatory Network (NEON) maintains an extensive collection of carabid specimens from across the United States, these primarily exist as physical collections, restricting widespread research access and large-scale analysis. To address these gaps, we present a multimodal dataset digitizing over 13,200 NEON carabids from 30 sites spanning the continental US and Hawaii through high-resolution imaging, enabling broader access and computational analysis. The dataset includes digitally measured elytra length and width of each specimen, establishing a foundation for automated trait extraction using AI. Validated against manual measurements, our digital trait extraction achieves sub-millimeter precision, ensuring reliability for ecological and computational studies. By addressing invertebrate under-representation in trait databases, this work supports AI-driven tools for automated species identification and trait-based research, fostering advancements in biodiversity monitoring and conservation.
Open World Scene Graph Generation using Vision Language Models
Jun 09, 2025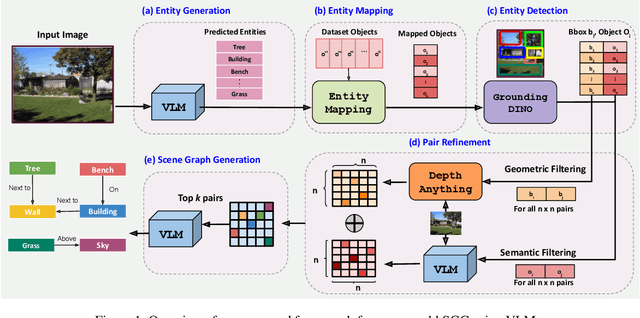
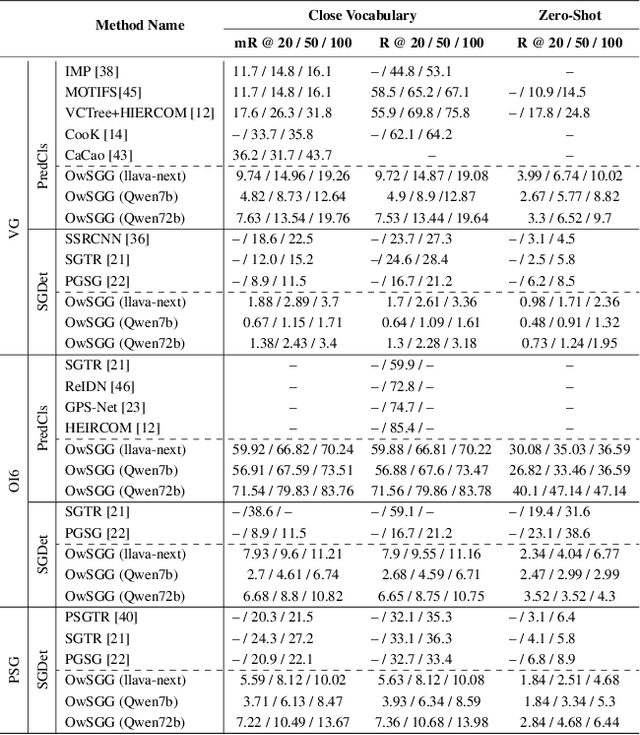
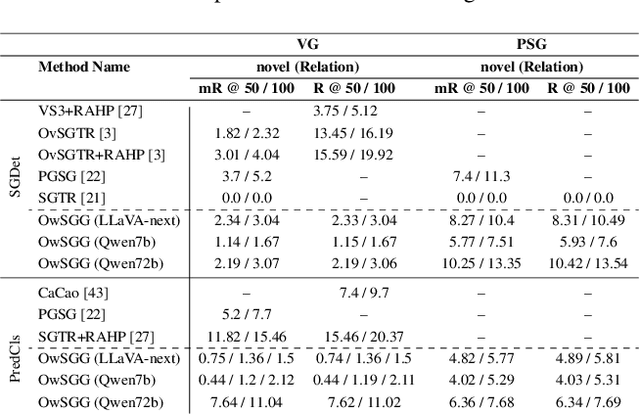
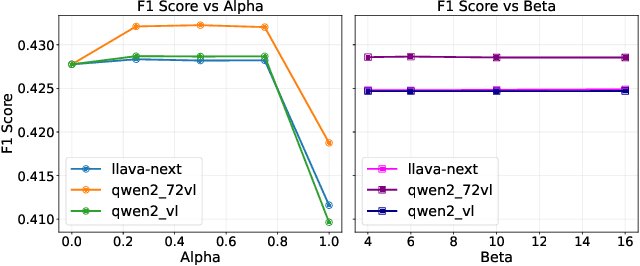
Abstract:Scene-Graph Generation (SGG) seeks to recognize objects in an image and distill their salient pairwise relationships. Most methods depend on dataset-specific supervision to learn the variety of interactions, restricting their usefulness in open-world settings, involving novel objects and/or relations. Even methods that leverage large Vision Language Models (VLMs) typically require benchmark-specific fine-tuning. We introduce Open-World SGG, a training-free, efficient, model-agnostic framework that taps directly into the pretrained knowledge of VLMs to produce scene graphs with zero additional learning. Casting SGG as a zero-shot structured-reasoning problem, our method combines multimodal prompting, embedding alignment, and a lightweight pair-refinement strategy, enabling inference over unseen object vocabularies and relation sets. To assess this setting, we formalize an Open-World evaluation protocol that measures performance when no SGG-specific data have been observed either in terms of objects and relations. Experiments on Visual Genome, Open Images V6, and the Panoptic Scene Graph (PSG) dataset demonstrate the capacity of pretrained VLMs to perform relational understanding without task-level training.
VLM4Bio: A Benchmark Dataset to Evaluate Pretrained Vision-Language Models for Trait Discovery from Biological Images
Aug 28, 2024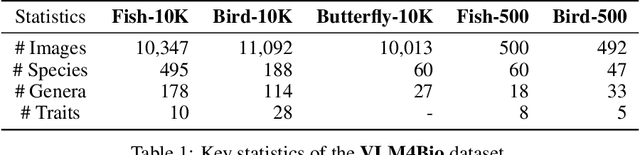
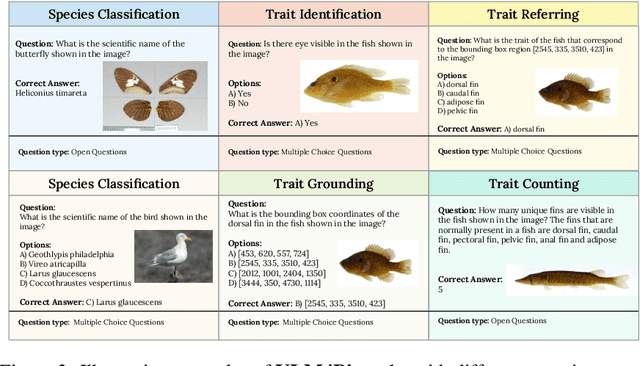

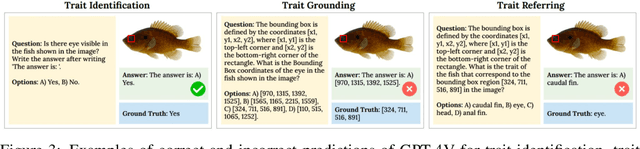
Abstract:Images are increasingly becoming the currency for documenting biodiversity on the planet, providing novel opportunities for accelerating scientific discoveries in the field of organismal biology, especially with the advent of large vision-language models (VLMs). We ask if pre-trained VLMs can aid scientists in answering a range of biologically relevant questions without any additional fine-tuning. In this paper, we evaluate the effectiveness of 12 state-of-the-art (SOTA) VLMs in the field of organismal biology using a novel dataset, VLM4Bio, consisting of 469K question-answer pairs involving 30K images from three groups of organisms: fishes, birds, and butterflies, covering five biologically relevant tasks. We also explore the effects of applying prompting techniques and tests for reasoning hallucination on the performance of VLMs, shedding new light on the capabilities of current SOTA VLMs in answering biologically relevant questions using images. The code and datasets for running all the analyses reported in this paper can be found at https://github.com/sammarfy/VLM4Bio.
Hierarchical Conditioning of Diffusion Models Using Tree-of-Life for Studying Species Evolution
Jul 31, 2024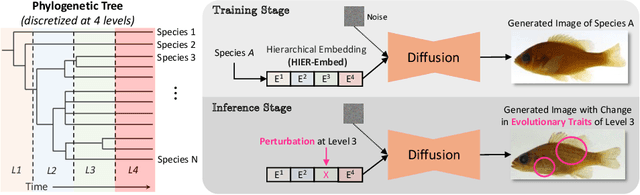
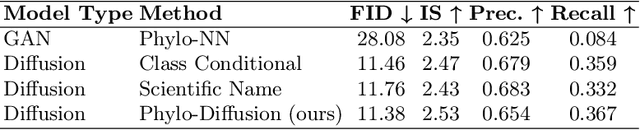
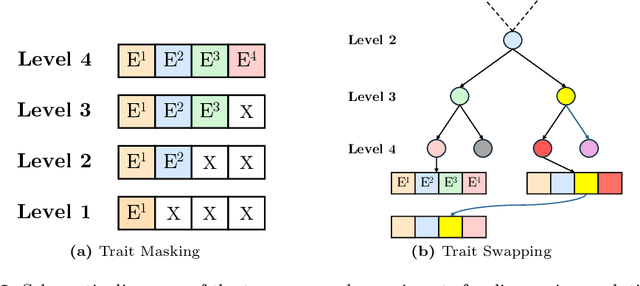

Abstract:A central problem in biology is to understand how organisms evolve and adapt to their environment by acquiring variations in the observable characteristics or traits of species across the tree of life. With the growing availability of large-scale image repositories in biology and recent advances in generative modeling, there is an opportunity to accelerate the discovery of evolutionary traits automatically from images. Toward this goal, we introduce Phylo-Diffusion, a novel framework for conditioning diffusion models with phylogenetic knowledge represented in the form of HIERarchical Embeddings (HIER-Embeds). We also propose two new experiments for perturbing the embedding space of Phylo-Diffusion: trait masking and trait swapping, inspired by counterpart experiments of gene knockout and gene editing/swapping. Our work represents a novel methodological advance in generative modeling to structure the embedding space of diffusion models using tree-based knowledge. Our work also opens a new chapter of research in evolutionary biology by using generative models to visualize evolutionary changes directly from images. We empirically demonstrate the usefulness of Phylo-Diffusion in capturing meaningful trait variations for fishes and birds, revealing novel insights about the biological mechanisms of their evolution.
Fish-Vista: A Multi-Purpose Dataset for Understanding & Identification of Traits from Images
Jul 10, 2024



Abstract:Fishes are integral to both ecological systems and economic sectors, and studying fish traits is crucial for understanding biodiversity patterns and macro-evolution trends. To enable the analysis of visual traits from fish images, we introduce the Fish-Visual Trait Analysis (Fish-Vista) dataset - a large, annotated collection of about 60K fish images spanning 1900 different species, supporting several challenging and biologically relevant tasks including species classification, trait identification, and trait segmentation. These images have been curated through a sophisticated data processing pipeline applied to a cumulative set of images obtained from various museum collections. Fish-Vista provides fine-grained labels of various visual traits present in each image. It also offers pixel-level annotations of 9 different traits for 2427 fish images, facilitating additional trait segmentation and localization tasks. The ultimate goal of Fish-Vista is to provide a clean, carefully curated, high-resolution dataset that can serve as a foundation for accelerating biological discoveries using advances in AI. Finally, we provide a comprehensive analysis of state-of-the-art deep learning techniques on Fish-Vista.
Discovering Novel Biological Traits From Images Using Phylogeny-Guided Neural Networks
Jun 05, 2023



Abstract:Discovering evolutionary traits that are heritable across species on the tree of life (also referred to as a phylogenetic tree) is of great interest to biologists to understand how organisms diversify and evolve. However, the measurement of traits is often a subjective and labor-intensive process, making trait discovery a highly label-scarce problem. We present a novel approach for discovering evolutionary traits directly from images without relying on trait labels. Our proposed approach, Phylo-NN, encodes the image of an organism into a sequence of quantized feature vectors -- or codes -- where different segments of the sequence capture evolutionary signals at varying ancestry levels in the phylogeny. We demonstrate the effectiveness of our approach in producing biologically meaningful results in a number of downstream tasks including species image generation and species-to-species image translation, using fish species as a target example.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge