James P. Balhoff
VLM4Bio: A Benchmark Dataset to Evaluate Pretrained Vision-Language Models for Trait Discovery from Biological Images
Aug 28, 2024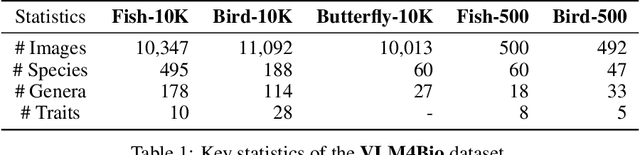
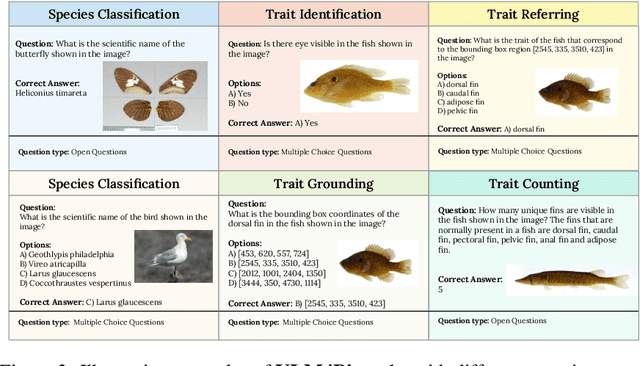

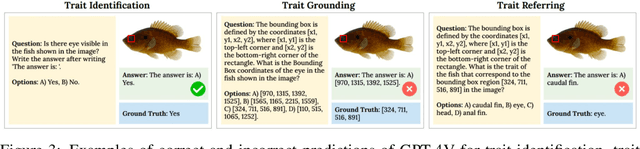
Abstract:Images are increasingly becoming the currency for documenting biodiversity on the planet, providing novel opportunities for accelerating scientific discoveries in the field of organismal biology, especially with the advent of large vision-language models (VLMs). We ask if pre-trained VLMs can aid scientists in answering a range of biologically relevant questions without any additional fine-tuning. In this paper, we evaluate the effectiveness of 12 state-of-the-art (SOTA) VLMs in the field of organismal biology using a novel dataset, VLM4Bio, consisting of 469K question-answer pairs involving 30K images from three groups of organisms: fishes, birds, and butterflies, covering five biologically relevant tasks. We also explore the effects of applying prompting techniques and tests for reasoning hallucination on the performance of VLMs, shedding new light on the capabilities of current SOTA VLMs in answering biologically relevant questions using images. The code and datasets for running all the analyses reported in this paper can be found at https://github.com/sammarfy/VLM4Bio.
Hierarchical Conditioning of Diffusion Models Using Tree-of-Life for Studying Species Evolution
Jul 31, 2024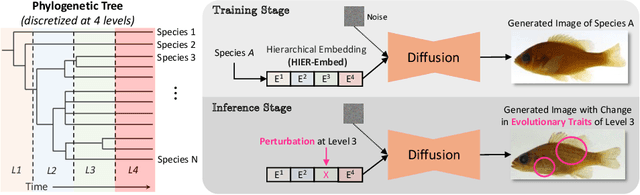
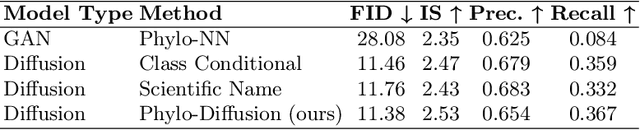
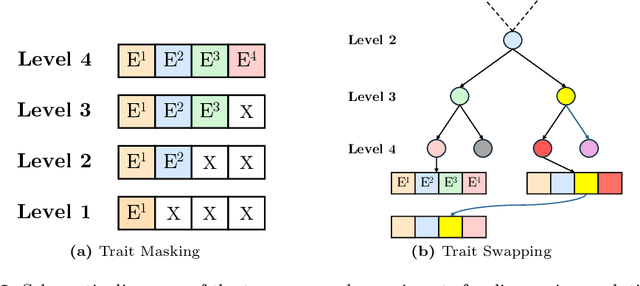

Abstract:A central problem in biology is to understand how organisms evolve and adapt to their environment by acquiring variations in the observable characteristics or traits of species across the tree of life. With the growing availability of large-scale image repositories in biology and recent advances in generative modeling, there is an opportunity to accelerate the discovery of evolutionary traits automatically from images. Toward this goal, we introduce Phylo-Diffusion, a novel framework for conditioning diffusion models with phylogenetic knowledge represented in the form of HIERarchical Embeddings (HIER-Embeds). We also propose two new experiments for perturbing the embedding space of Phylo-Diffusion: trait masking and trait swapping, inspired by counterpart experiments of gene knockout and gene editing/swapping. Our work represents a novel methodological advance in generative modeling to structure the embedding space of diffusion models using tree-based knowledge. Our work also opens a new chapter of research in evolutionary biology by using generative models to visualize evolutionary changes directly from images. We empirically demonstrate the usefulness of Phylo-Diffusion in capturing meaningful trait variations for fishes and birds, revealing novel insights about the biological mechanisms of their evolution.
Discovering Novel Biological Traits From Images Using Phylogeny-Guided Neural Networks
Jun 05, 2023



Abstract:Discovering evolutionary traits that are heritable across species on the tree of life (also referred to as a phylogenetic tree) is of great interest to biologists to understand how organisms diversify and evolve. However, the measurement of traits is often a subjective and labor-intensive process, making trait discovery a highly label-scarce problem. We present a novel approach for discovering evolutionary traits directly from images without relying on trait labels. Our proposed approach, Phylo-NN, encodes the image of an organism into a sequence of quantized feature vectors -- or codes -- where different segments of the sequence capture evolutionary signals at varying ancestry levels in the phylogeny. We demonstrate the effectiveness of our approach in producing biologically meaningful results in a number of downstream tasks including species image generation and species-to-species image translation, using fish species as a target example.
Presence-absence reasoning for evolutionary phenotypes
Oct 14, 2014Abstract:Nearly invariably, phenotypes are reported in the scientific literature in meticulous detail, utilizing the full expressivity of natural language. Often it is particularly these detailed observations (facts) that are of interest, and thus specific to the research questions that motivated observing and reporting them. However, research aiming to synthesize or integrate phenotype data across many studies or even fields is often faced with the need to abstract from detailed observations so as to construct phenotypic concepts that are common across many datasets rather than specific to a few. Yet, observations or facts that would fall under such abstracted concepts are typically not directly asserted by the original authors, usually because they are "obvious" according to common domain knowledge, and thus asserting them would be deemed redundant by anyone with sufficient domain knowledge. For example, a phenotype describing the length of a manual digit for an organism implicitly means that the organism must have had a hand, and thus a forelimb; the presence or absence of a forelimb may have supporting data across a far wider range of taxa than the length of a particular manual digit. Here we describe how within the Phenoscape project we use a pipeline of OWL axiom generation and reasoning steps to infer taxon-specific presence/absence of anatomical entities from anatomical phenotypes. Although presence/absence is all but one, and a seemingly simple way to abstract phenotypes across data sources, it can nonetheless be powerful for linking genotype to phenotype, and it is particularly relevant for constructing synthetic morphological supermatrices for comparative analysis; in fact presence/absence is one of the prevailing character observation types in published character matrices.
* 4 pages. Peer-reviewed submission presented to the Bio-ontologies SIG Phenotype Day at ISMB 2014, Boston, Mass. http://phenoday2014.bio-lark.org/pdf/11.pdf
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge