M. Maruf
Open World Scene Graph Generation using Vision Language Models
Jun 09, 2025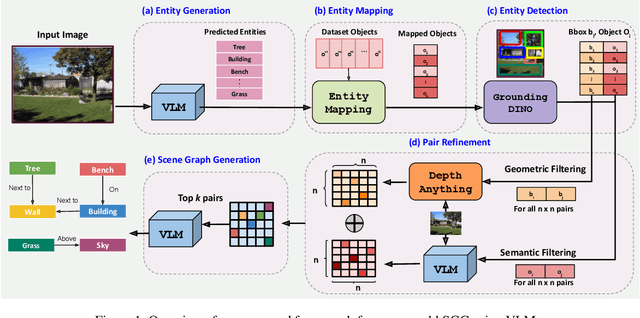
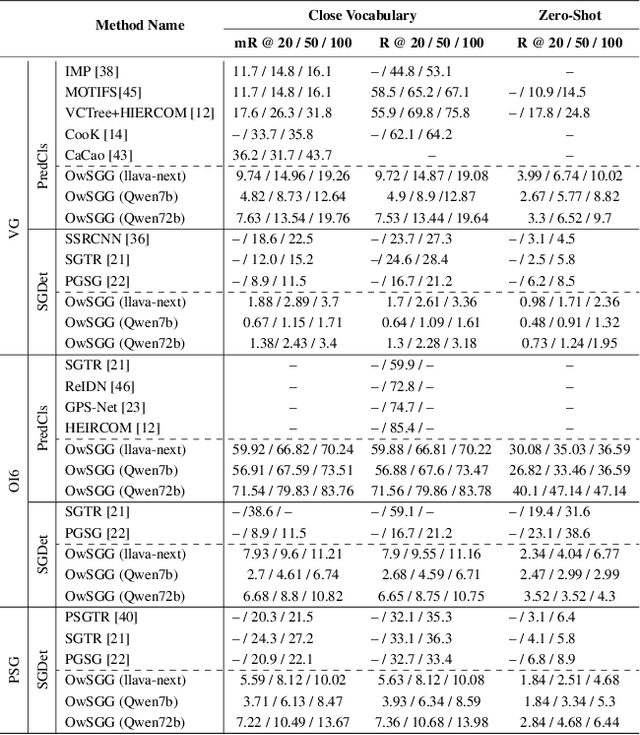
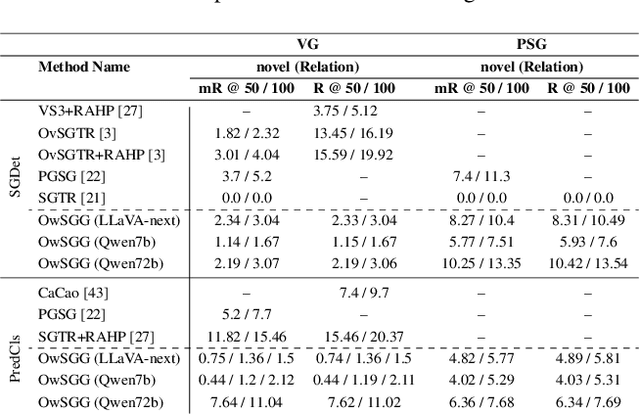
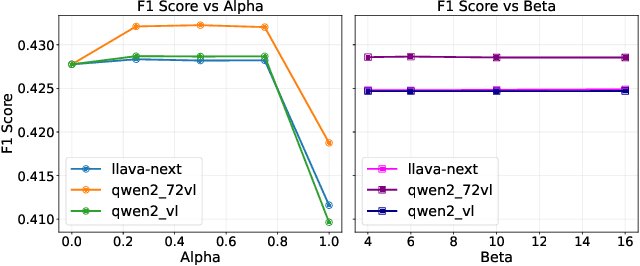
Abstract:Scene-Graph Generation (SGG) seeks to recognize objects in an image and distill their salient pairwise relationships. Most methods depend on dataset-specific supervision to learn the variety of interactions, restricting their usefulness in open-world settings, involving novel objects and/or relations. Even methods that leverage large Vision Language Models (VLMs) typically require benchmark-specific fine-tuning. We introduce Open-World SGG, a training-free, efficient, model-agnostic framework that taps directly into the pretrained knowledge of VLMs to produce scene graphs with zero additional learning. Casting SGG as a zero-shot structured-reasoning problem, our method combines multimodal prompting, embedding alignment, and a lightweight pair-refinement strategy, enabling inference over unseen object vocabularies and relation sets. To assess this setting, we formalize an Open-World evaluation protocol that measures performance when no SGG-specific data have been observed either in terms of objects and relations. Experiments on Visual Genome, Open Images V6, and the Panoptic Scene Graph (PSG) dataset demonstrate the capacity of pretrained VLMs to perform relational understanding without task-level training.
What Do You See in Common? Learning Hierarchical Prototypes over Tree-of-Life to Discover Evolutionary Traits
Sep 03, 2024

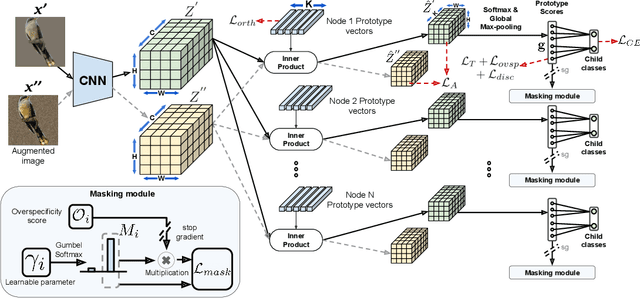

Abstract:A grand challenge in biology is to discover evolutionary traits - features of organisms common to a group of species with a shared ancestor in the tree of life (also referred to as phylogenetic tree). With the growing availability of image repositories in biology, there is a tremendous opportunity to discover evolutionary traits directly from images in the form of a hierarchy of prototypes. However, current prototype-based methods are mostly designed to operate over a flat structure of classes and face several challenges in discovering hierarchical prototypes, including the issue of learning over-specific features at internal nodes. To overcome these challenges, we introduce the framework of Hierarchy aligned Commonality through Prototypical Networks (HComP-Net). We empirically show that HComP-Net learns prototypes that are accurate, semantically consistent, and generalizable to unseen species in comparison to baselines on birds, butterflies, and fishes datasets. The code and datasets are available at https://github.com/Imageomics/HComPNet.
VLM4Bio: A Benchmark Dataset to Evaluate Pretrained Vision-Language Models for Trait Discovery from Biological Images
Aug 28, 2024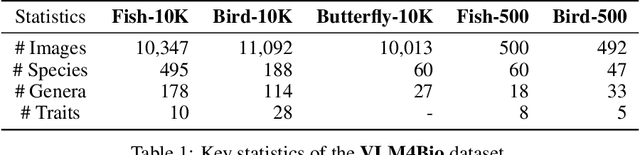
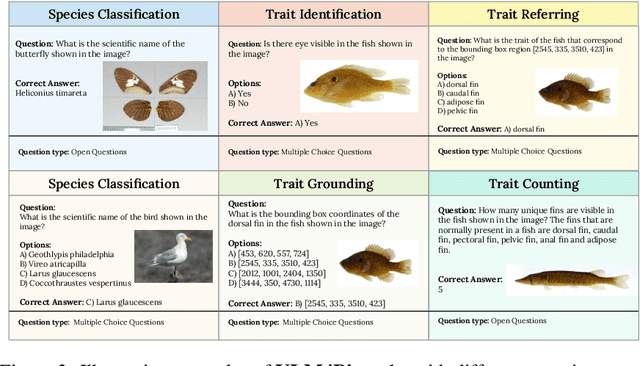

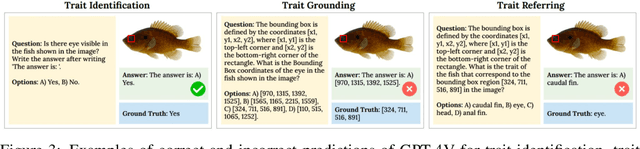
Abstract:Images are increasingly becoming the currency for documenting biodiversity on the planet, providing novel opportunities for accelerating scientific discoveries in the field of organismal biology, especially with the advent of large vision-language models (VLMs). We ask if pre-trained VLMs can aid scientists in answering a range of biologically relevant questions without any additional fine-tuning. In this paper, we evaluate the effectiveness of 12 state-of-the-art (SOTA) VLMs in the field of organismal biology using a novel dataset, VLM4Bio, consisting of 469K question-answer pairs involving 30K images from three groups of organisms: fishes, birds, and butterflies, covering five biologically relevant tasks. We also explore the effects of applying prompting techniques and tests for reasoning hallucination on the performance of VLMs, shedding new light on the capabilities of current SOTA VLMs in answering biologically relevant questions using images. The code and datasets for running all the analyses reported in this paper can be found at https://github.com/sammarfy/VLM4Bio.
Hierarchical Conditioning of Diffusion Models Using Tree-of-Life for Studying Species Evolution
Jul 31, 2024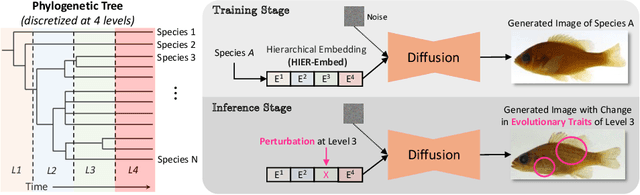
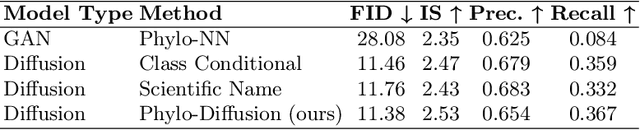
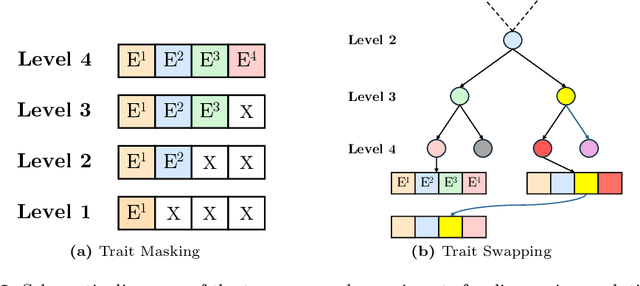

Abstract:A central problem in biology is to understand how organisms evolve and adapt to their environment by acquiring variations in the observable characteristics or traits of species across the tree of life. With the growing availability of large-scale image repositories in biology and recent advances in generative modeling, there is an opportunity to accelerate the discovery of evolutionary traits automatically from images. Toward this goal, we introduce Phylo-Diffusion, a novel framework for conditioning diffusion models with phylogenetic knowledge represented in the form of HIERarchical Embeddings (HIER-Embeds). We also propose two new experiments for perturbing the embedding space of Phylo-Diffusion: trait masking and trait swapping, inspired by counterpart experiments of gene knockout and gene editing/swapping. Our work represents a novel methodological advance in generative modeling to structure the embedding space of diffusion models using tree-based knowledge. Our work also opens a new chapter of research in evolutionary biology by using generative models to visualize evolutionary changes directly from images. We empirically demonstrate the usefulness of Phylo-Diffusion in capturing meaningful trait variations for fishes and birds, revealing novel insights about the biological mechanisms of their evolution.
Fish-Vista: A Multi-Purpose Dataset for Understanding & Identification of Traits from Images
Jul 10, 2024



Abstract:Fishes are integral to both ecological systems and economic sectors, and studying fish traits is crucial for understanding biodiversity patterns and macro-evolution trends. To enable the analysis of visual traits from fish images, we introduce the Fish-Visual Trait Analysis (Fish-Vista) dataset - a large, annotated collection of about 60K fish images spanning 1900 different species, supporting several challenging and biologically relevant tasks including species classification, trait identification, and trait segmentation. These images have been curated through a sophisticated data processing pipeline applied to a cumulative set of images obtained from various museum collections. Fish-Vista provides fine-grained labels of various visual traits present in each image. It also offers pixel-level annotations of 9 different traits for 2427 fish images, facilitating additional trait segmentation and localization tasks. The ultimate goal of Fish-Vista is to provide a clean, carefully curated, high-resolution dataset that can serve as a foundation for accelerating biological discoveries using advances in AI. Finally, we provide a comprehensive analysis of state-of-the-art deep learning techniques on Fish-Vista.
Beyond Discriminative Regions: Saliency Maps as Alternatives to CAMs for Weakly Supervised Semantic Segmentation
Aug 21, 2023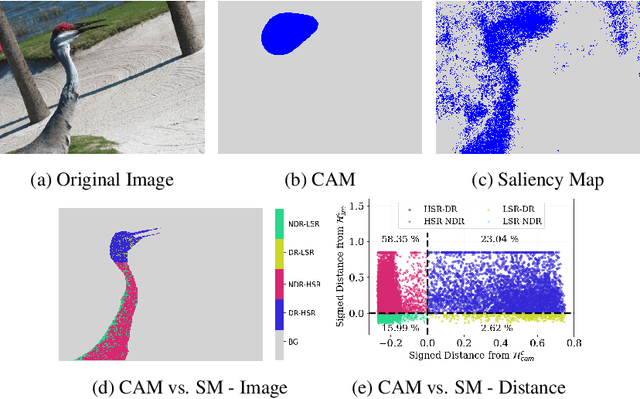
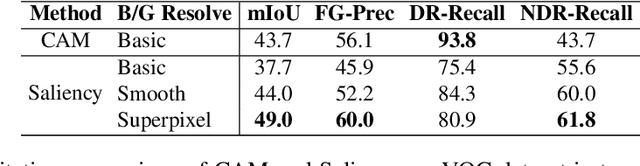
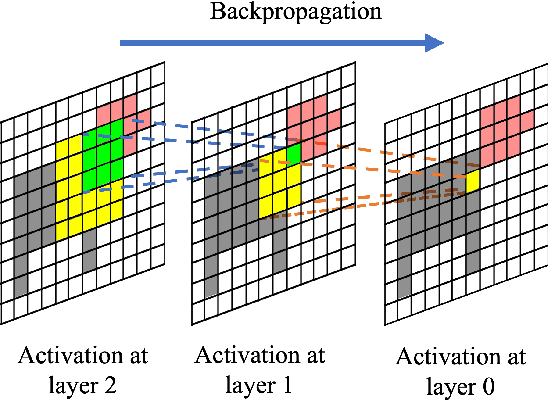
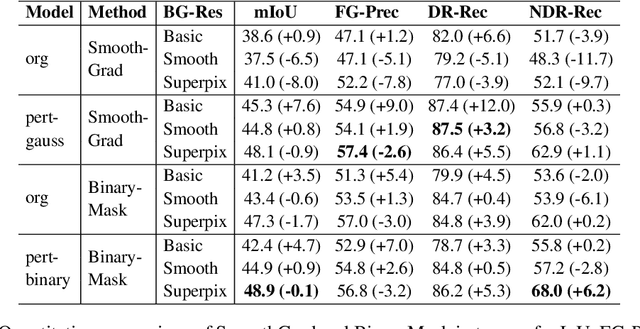
Abstract:In recent years, several Weakly Supervised Semantic Segmentation (WS3) methods have been proposed that use class activation maps (CAMs) generated by a classifier to produce pseudo-ground truths for training segmentation models. While CAMs are good at highlighting discriminative regions (DR) of an image, they are known to disregard regions of the object that do not contribute to the classifier's prediction, termed non-discriminative regions (NDR). In contrast, attribution methods such as saliency maps provide an alternative approach for assigning a score to every pixel based on its contribution to the classification prediction. This paper provides a comprehensive comparison between saliencies and CAMs for WS3. Our study includes multiple perspectives on understanding their similarities and dissimilarities. Moreover, we provide new evaluation metrics that perform a comprehensive assessment of WS3 performance of alternative methods w.r.t. CAMs. We demonstrate the effectiveness of saliencies in addressing the limitation of CAMs through our empirical studies on benchmark datasets. Furthermore, we propose random cropping as a stochastic aggregation technique that improves the performance of saliency, making it a strong alternative to CAM for WS3.
Learning Compact Representations of Neural Networks using DiscriminAtive Masking (DAM)
Oct 01, 2021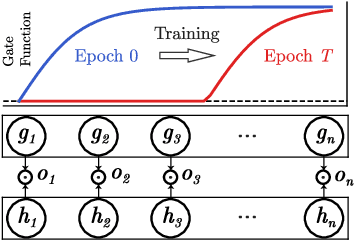

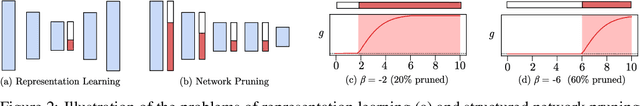

Abstract:A central goal in deep learning is to learn compact representations of features at every layer of a neural network, which is useful for both unsupervised representation learning and structured network pruning. While there is a growing body of work in structured pruning, current state-of-the-art methods suffer from two key limitations: (i) instability during training, and (ii) need for an additional step of fine-tuning, which is resource-intensive. At the core of these limitations is the lack of a systematic approach that jointly prunes and refines weights during training in a single stage, and does not require any fine-tuning upon convergence to achieve state-of-the-art performance. We present a novel single-stage structured pruning method termed DiscriminAtive Masking (DAM). The key intuition behind DAM is to discriminatively prefer some of the neurons to be refined during the training process, while gradually masking out other neurons. We show that our proposed DAM approach has remarkably good performance over various applications, including dimensionality reduction, recommendation system, graph representation learning, and structured pruning for image classification. We also theoretically show that the learning objective of DAM is directly related to minimizing the L0 norm of the masking layer.
PID-GAN: A GAN Framework based on a Physics-informed Discriminator for Uncertainty Quantification with Physics
Jun 06, 2021



Abstract:As applications of deep learning (DL) continue to seep into critical scientific use-cases, the importance of performing uncertainty quantification (UQ) with DL has become more pressing than ever before. In scientific applications, it is also important to inform the learning of DL models with knowledge of physics of the problem to produce physically consistent and generalized solutions. This is referred to as the emerging field of physics-informed deep learning (PIDL). We consider the problem of developing PIDL formulations that can also perform UQ. To this end, we propose a novel physics-informed GAN architecture, termed PID-GAN, where the knowledge of physics is used to inform the learning of both the generator and discriminator models, making ample use of unlabeled data instances. We show that our proposed PID-GAN framework does not suffer from imbalance of generator gradients from multiple loss terms as compared to state-of-the-art. We also empirically demonstrate the efficacy of our proposed framework on a variety of case studies involving benchmark physics-based PDEs as well as imperfect physics. All the code and datasets used in this study have been made available on this link : https://github.com/arkadaw9/PID-GAN.
Beyond Observed Connections : Link Injection
Sep 02, 2020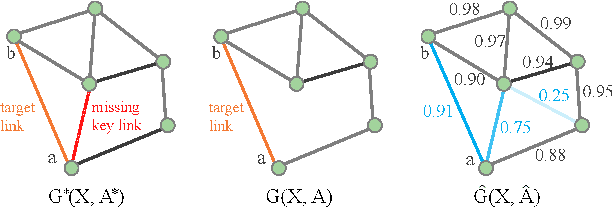
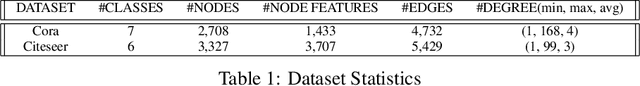
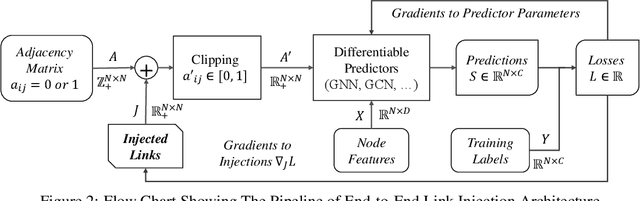
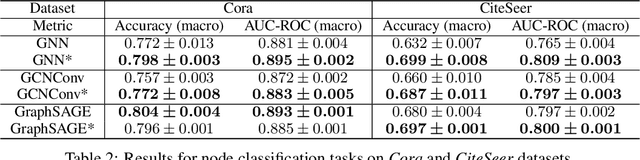
Abstract:In this paper, we proposed the \textit{link injection}, a novel method that helps any differentiable graph machine learning models to go beyond observed connections from the input data in an end-to-end learning fashion. It finds out (weak) connections in favor of the current task that is not present in the input data via a parametric link injection layer. We evaluate our method on both node classification and link prediction tasks using a series of state-of-the-art graph convolution networks. Results show that the link injection helps a variety of models to achieve better performances on both applications. Further empirical analysis shows a great potential of this method in efficiently exploiting unseen connections from the injected links.
Maximizing Cohesion and Separation in Graph Representation Learning: A Distance-aware Negative Sampling Approach
Jul 02, 2020



Abstract:The objective of unsupervised graph representation learning (GRL) is to learn a low-dimensional space of node embeddings that reflect the structure of a given unlabeled graph. Existing algorithms for this task rely on negative sampling objectives that maximize the similarity in node embeddings at nearby nodes (referred to as "cohesion") by maintaining positive and negative corpus of node pairs. While positive samples are drawn from node pairs that co-occur in short random walks, conventional approaches construct negative corpus by uniformly sampling random pairs, thus ignoring valuable information about structural dissimilarity among distant node pairs (referred to as "separation"). In this paper, we present a novel Distance-aware Negative Sampling (DNS) which maximizes the separation of distant node-pairs while maximizing cohesion at nearby node-pairs by setting the negative sampling probability proportional to the pair-wise shortest distances. Our approach can be used in conjunction with any GRL algorithm and we demonstrate the efficacy of our approach over baseline negative sampling methods over downstream node classification tasks on a number of benchmark datasets and GRL algorithms. All our codes and datasets are available at \url{https://github.com/Distance-awareNS/DNS/}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge