Anuj Karpatne
A continental-scale dataset of ground beetles with high-resolution images and validated morphological trait measurements
Jan 14, 2026Abstract:Despite the ecological significance of invertebrates, global trait databases remain heavily biased toward vertebrates and plants, limiting comprehensive ecological analyses of high-diversity groups like ground beetles. Ground beetles (Coleoptera: Carabidae) serve as critical bioindicators of ecosystem health, providing valuable insights into biodiversity shifts driven by environmental changes. While the National Ecological Observatory Network (NEON) maintains an extensive collection of carabid specimens from across the United States, these primarily exist as physical collections, restricting widespread research access and large-scale analysis. To address these gaps, we present a multimodal dataset digitizing over 13,200 NEON carabids from 30 sites spanning the continental US and Hawaii through high-resolution imaging, enabling broader access and computational analysis. The dataset includes digitally measured elytra length and width of each specimen, establishing a foundation for automated trait extraction using AI. Validated against manual measurements, our digital trait extraction achieves sub-millimeter precision, ensuring reliability for ecological and computational studies. By addressing invertebrate under-representation in trait databases, this work supports AI-driven tools for automated species identification and trait-based research, fostering advancements in biodiversity monitoring and conservation.
Open World Scene Graph Generation using Vision Language Models
Jun 09, 2025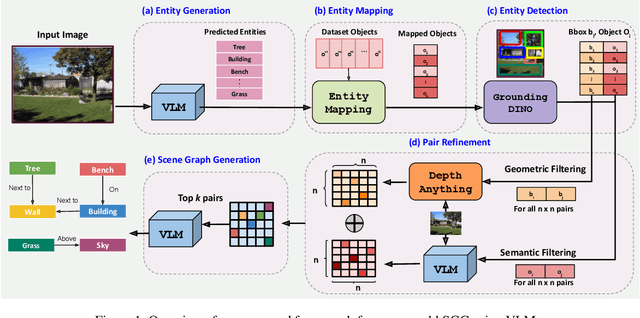
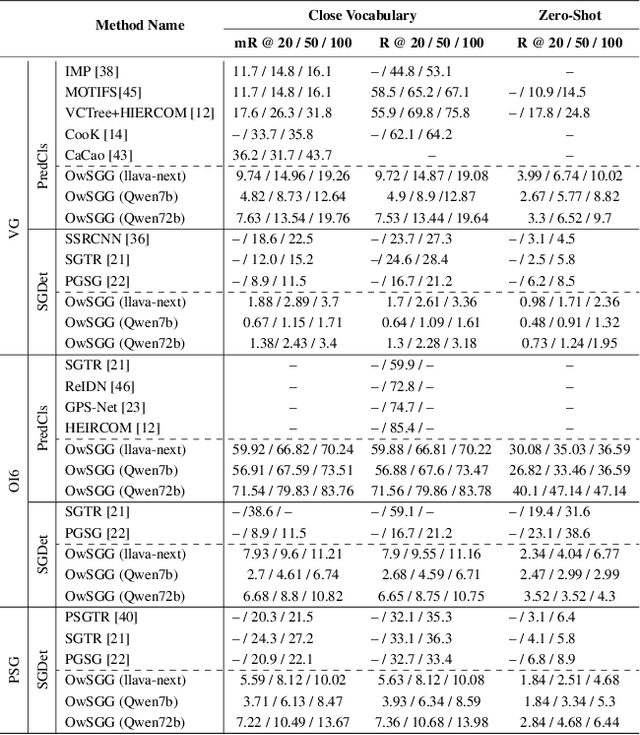
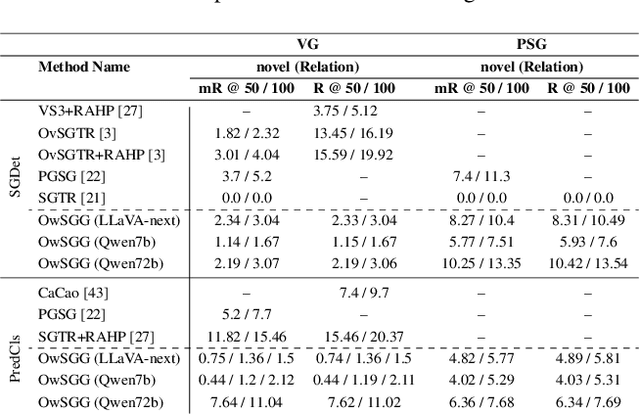
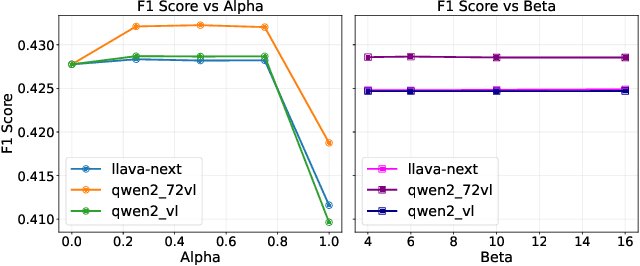
Abstract:Scene-Graph Generation (SGG) seeks to recognize objects in an image and distill their salient pairwise relationships. Most methods depend on dataset-specific supervision to learn the variety of interactions, restricting their usefulness in open-world settings, involving novel objects and/or relations. Even methods that leverage large Vision Language Models (VLMs) typically require benchmark-specific fine-tuning. We introduce Open-World SGG, a training-free, efficient, model-agnostic framework that taps directly into the pretrained knowledge of VLMs to produce scene graphs with zero additional learning. Casting SGG as a zero-shot structured-reasoning problem, our method combines multimodal prompting, embedding alignment, and a lightweight pair-refinement strategy, enabling inference over unseen object vocabularies and relation sets. To assess this setting, we formalize an Open-World evaluation protocol that measures performance when no SGG-specific data have been observed either in terms of objects and relations. Experiments on Visual Genome, Open Images V6, and the Panoptic Scene Graph (PSG) dataset demonstrate the capacity of pretrained VLMs to perform relational understanding without task-level training.
Prompt-CAM: A Simpler Interpretable Transformer for Fine-Grained Analysis
Jan 16, 2025



Abstract:We present a simple usage of pre-trained Vision Transformers (ViTs) for fine-grained analysis, aiming to identify and localize the traits that distinguish visually similar categories, such as different bird species or dog breeds. Pre-trained ViTs such as DINO have shown remarkable capabilities to extract localized, informative features. However, using saliency maps like Grad-CAM can hardly point out the traits: they often locate the whole object by a blurred, coarse heatmap, not traits. We propose a novel approach Prompt Class Attention Map (Prompt-CAM) to the rescue. Prompt-CAM learns class-specific prompts to a pre-trained ViT and uses the corresponding outputs for classification. To classify an image correctly, the true-class prompt must attend to the unique image patches not seen in other classes' images, i.e., traits. As such, the true class's multi-head attention maps reveal traits and their locations. Implementation-wise, Prompt-CAM is almost a free lunch by simply modifying the prediction head of Visual Prompt Tuning (VPT). This makes Prompt-CAM fairly easy to train and apply, sharply contrasting other interpretable methods that design specific models and training processes. It is even simpler than the recently published INterpretable TRansformer (INTR), whose encoder-decoder architecture prevents it from leveraging pre-trained ViTs. Extensive empirical studies on a dozen datasets from various domains (e.g., birds, fishes, insects, fungi, flowers, food, and cars) validate Prompt-CAM superior interpretation capability.
Static Segmentation by Tracking: A Frustratingly Label-Efficient Approach to Fine-Grained Segmentation
Jan 12, 2025



Abstract:We study image segmentation in the biological domain, particularly trait and part segmentation from specimen images (e.g., butterfly wing stripes or beetle body parts). This is a crucial, fine-grained task that aids in understanding the biology of organisms. The conventional approach involves hand-labeling masks, often for hundreds of images per species, and training a segmentation model to generalize these labels to other images, which can be exceedingly laborious. We present a label-efficient method named Static Segmentation by Tracking (SST). SST is built upon the insight: while specimens of the same species have inherent variations, the traits and parts we aim to segment show up consistently. This motivates us to concatenate specimen images into a ``pseudo-video'' and reframe trait and part segmentation as a tracking problem. Concretely, SST generates masks for unlabeled images by propagating annotated or predicted masks from the ``pseudo-preceding'' images. Powered by Segment Anything Model 2 (SAM~2) initially developed for video segmentation, we show that SST can achieve high-quality trait and part segmentation with merely one labeled image per species -- a breakthrough for analyzing specimen images. We further develop a cycle-consistent loss to fine-tune the model, again using one labeled image. Additionally, we highlight the broader potential of SST, including one-shot instance segmentation on images taken in the wild and trait-based image retrieval.
A Unified Framework for Forward and Inverse Problems in Subsurface Imaging using Latent Space Translations
Oct 15, 2024Abstract:In subsurface imaging, learning the mapping from velocity maps to seismic waveforms (forward problem) and waveforms to velocity (inverse problem) is important for several applications. While traditional techniques for solving forward and inverse problems are computationally prohibitive, there is a growing interest in leveraging recent advances in deep learning to learn the mapping between velocity maps and seismic waveform images directly from data. Despite the variety of architectures explored in previous works, several open questions still remain unanswered such as the effect of latent space sizes, the importance of manifold learning, the complexity of translation models, and the value of jointly solving forward and inverse problems. We propose a unified framework to systematically characterize prior research in this area termed the Generalized Forward-Inverse (GFI) framework, building on the assumption of manifolds and latent space translations. We show that GFI encompasses previous works in deep learning for subsurface imaging, which can be viewed as specific instantiations of GFI. We also propose two new model architectures within the framework of GFI: Latent U-Net and Invertible X-Net, leveraging the power of U-Nets for domain translation and the ability of IU-Nets to simultaneously learn forward and inverse translations, respectively. We show that our proposed models achieve state-of-the-art (SOTA) performance for forward and inverse problems on a wide range of synthetic datasets, and also investigate their zero-shot effectiveness on two real-world-like datasets.
What Do You See in Common? Learning Hierarchical Prototypes over Tree-of-Life to Discover Evolutionary Traits
Sep 03, 2024

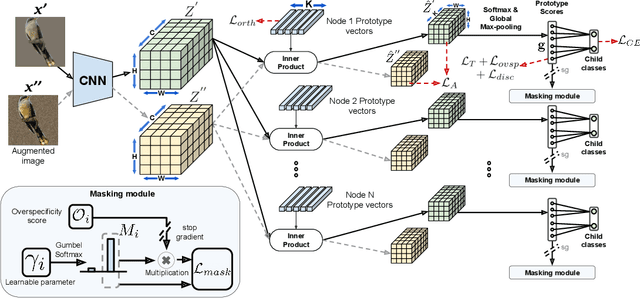

Abstract:A grand challenge in biology is to discover evolutionary traits - features of organisms common to a group of species with a shared ancestor in the tree of life (also referred to as phylogenetic tree). With the growing availability of image repositories in biology, there is a tremendous opportunity to discover evolutionary traits directly from images in the form of a hierarchy of prototypes. However, current prototype-based methods are mostly designed to operate over a flat structure of classes and face several challenges in discovering hierarchical prototypes, including the issue of learning over-specific features at internal nodes. To overcome these challenges, we introduce the framework of Hierarchy aligned Commonality through Prototypical Networks (HComP-Net). We empirically show that HComP-Net learns prototypes that are accurate, semantically consistent, and generalizable to unseen species in comparison to baselines on birds, butterflies, and fishes datasets. The code and datasets are available at https://github.com/Imageomics/HComPNet.
VLM4Bio: A Benchmark Dataset to Evaluate Pretrained Vision-Language Models for Trait Discovery from Biological Images
Aug 28, 2024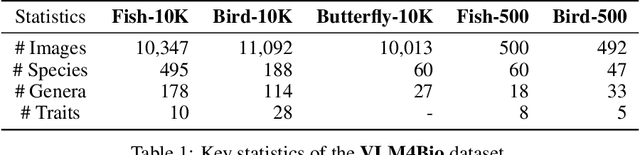
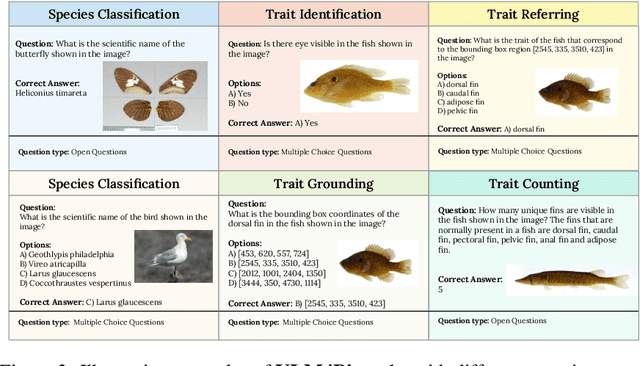

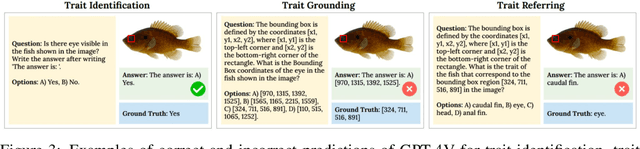
Abstract:Images are increasingly becoming the currency for documenting biodiversity on the planet, providing novel opportunities for accelerating scientific discoveries in the field of organismal biology, especially with the advent of large vision-language models (VLMs). We ask if pre-trained VLMs can aid scientists in answering a range of biologically relevant questions without any additional fine-tuning. In this paper, we evaluate the effectiveness of 12 state-of-the-art (SOTA) VLMs in the field of organismal biology using a novel dataset, VLM4Bio, consisting of 469K question-answer pairs involving 30K images from three groups of organisms: fishes, birds, and butterflies, covering five biologically relevant tasks. We also explore the effects of applying prompting techniques and tests for reasoning hallucination on the performance of VLMs, shedding new light on the capabilities of current SOTA VLMs in answering biologically relevant questions using images. The code and datasets for running all the analyses reported in this paper can be found at https://github.com/sammarfy/VLM4Bio.
Hierarchical Conditioning of Diffusion Models Using Tree-of-Life for Studying Species Evolution
Jul 31, 2024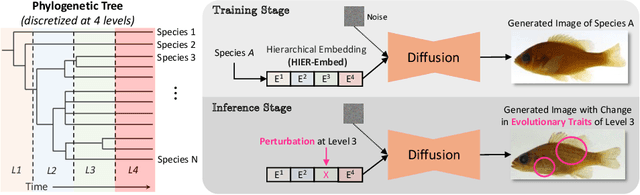
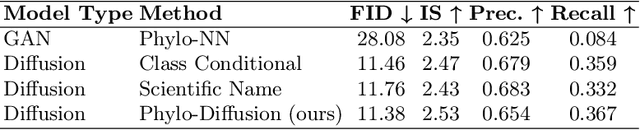
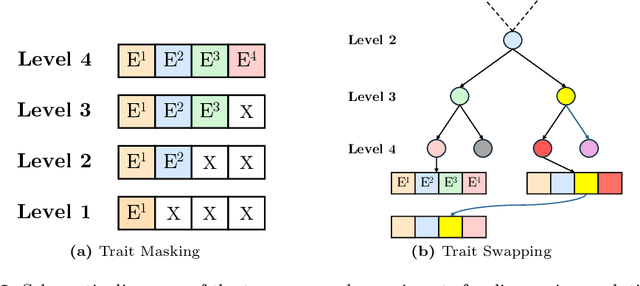

Abstract:A central problem in biology is to understand how organisms evolve and adapt to their environment by acquiring variations in the observable characteristics or traits of species across the tree of life. With the growing availability of large-scale image repositories in biology and recent advances in generative modeling, there is an opportunity to accelerate the discovery of evolutionary traits automatically from images. Toward this goal, we introduce Phylo-Diffusion, a novel framework for conditioning diffusion models with phylogenetic knowledge represented in the form of HIERarchical Embeddings (HIER-Embeds). We also propose two new experiments for perturbing the embedding space of Phylo-Diffusion: trait masking and trait swapping, inspired by counterpart experiments of gene knockout and gene editing/swapping. Our work represents a novel methodological advance in generative modeling to structure the embedding space of diffusion models using tree-based knowledge. Our work also opens a new chapter of research in evolutionary biology by using generative models to visualize evolutionary changes directly from images. We empirically demonstrate the usefulness of Phylo-Diffusion in capturing meaningful trait variations for fishes and birds, revealing novel insights about the biological mechanisms of their evolution.
Fish-Vista: A Multi-Purpose Dataset for Understanding & Identification of Traits from Images
Jul 10, 2024



Abstract:Fishes are integral to both ecological systems and economic sectors, and studying fish traits is crucial for understanding biodiversity patterns and macro-evolution trends. To enable the analysis of visual traits from fish images, we introduce the Fish-Visual Trait Analysis (Fish-Vista) dataset - a large, annotated collection of about 60K fish images spanning 1900 different species, supporting several challenging and biologically relevant tasks including species classification, trait identification, and trait segmentation. These images have been curated through a sophisticated data processing pipeline applied to a cumulative set of images obtained from various museum collections. Fish-Vista provides fine-grained labels of various visual traits present in each image. It also offers pixel-level annotations of 9 different traits for 2427 fish images, facilitating additional trait segmentation and localization tasks. The ultimate goal of Fish-Vista is to provide a clean, carefully curated, high-resolution dataset that can serve as a foundation for accelerating biological discoveries using advances in AI. Finally, we provide a comprehensive analysis of state-of-the-art deep learning techniques on Fish-Vista.
Knowledge-guided Machine Learning: Current Trends and Future Prospects
Mar 24, 2024



Abstract:This paper presents an overview of scientific modeling and discusses the complementary strengths and weaknesses of ML methods for scientific modeling in comparison to process-based models. It also provides an introduction to the current state of research in the emerging field of scientific knowledge-guided machine learning (KGML) that aims to use both scientific knowledge and data in ML frameworks to achieve better generalizability, scientific consistency, and explainability of results. We discuss different facets of KGML research in terms of the type of scientific knowledge used, the form of knowledge-ML integration explored, and the method for incorporating scientific knowledge in ML. We also discuss some of the common categories of use cases in environmental sciences where KGML methods are being developed, using illustrative examples in each category.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge