Lemeng Wang
AVA-Bench: Atomic Visual Ability Benchmark for Vision Foundation Models
Jun 10, 2025



Abstract:The rise of vision foundation models (VFMs) calls for systematic evaluation. A common approach pairs VFMs with large language models (LLMs) as general-purpose heads, followed by evaluation on broad Visual Question Answering (VQA) benchmarks. However, this protocol has two key blind spots: (i) the instruction tuning data may not align with VQA test distributions, meaning a wrong prediction can stem from such data mismatch rather than a VFM' visual shortcomings; (ii) VQA benchmarks often require multiple visual abilities, making it hard to tell whether errors stem from lacking all required abilities or just a single critical one. To address these gaps, we introduce AVA-Bench, the first benchmark that explicitly disentangles 14 Atomic Visual Abilities (AVAs) -- foundational skills like localization, depth estimation, and spatial understanding that collectively support complex visual reasoning tasks. By decoupling AVAs and matching training and test distributions within each, AVA-Bench pinpoints exactly where a VFM excels or falters. Applying AVA-Bench to leading VFMs thus reveals distinctive "ability fingerprints," turning VFM selection from educated guesswork into principled engineering. Notably, we find that a 0.5B LLM yields similar VFM rankings as a 7B LLM while cutting GPU hours by 8x, enabling more efficient evaluation. By offering a comprehensive and transparent benchmark, we hope AVA-Bench lays the foundation for the next generation of VFMs.
Static Segmentation by Tracking: A Frustratingly Label-Efficient Approach to Fine-Grained Segmentation
Jan 12, 2025



Abstract:We study image segmentation in the biological domain, particularly trait and part segmentation from specimen images (e.g., butterfly wing stripes or beetle body parts). This is a crucial, fine-grained task that aids in understanding the biology of organisms. The conventional approach involves hand-labeling masks, often for hundreds of images per species, and training a segmentation model to generalize these labels to other images, which can be exceedingly laborious. We present a label-efficient method named Static Segmentation by Tracking (SST). SST is built upon the insight: while specimens of the same species have inherent variations, the traits and parts we aim to segment show up consistently. This motivates us to concatenate specimen images into a ``pseudo-video'' and reframe trait and part segmentation as a tracking problem. Concretely, SST generates masks for unlabeled images by propagating annotated or predicted masks from the ``pseudo-preceding'' images. Powered by Segment Anything Model 2 (SAM~2) initially developed for video segmentation, we show that SST can achieve high-quality trait and part segmentation with merely one labeled image per species -- a breakthrough for analyzing specimen images. We further develop a cycle-consistent loss to fine-tune the model, again using one labeled image. Additionally, we highlight the broader potential of SST, including one-shot instance segmentation on images taken in the wild and trait-based image retrieval.
CompBench: A Comparative Reasoning Benchmark for Multimodal LLMs
Jul 23, 2024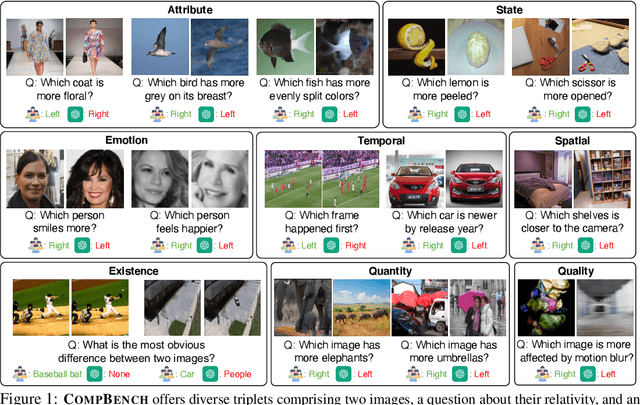
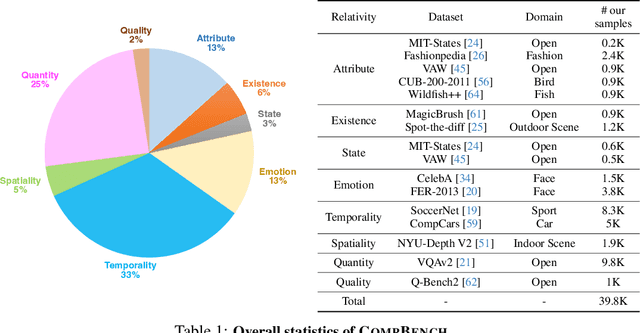
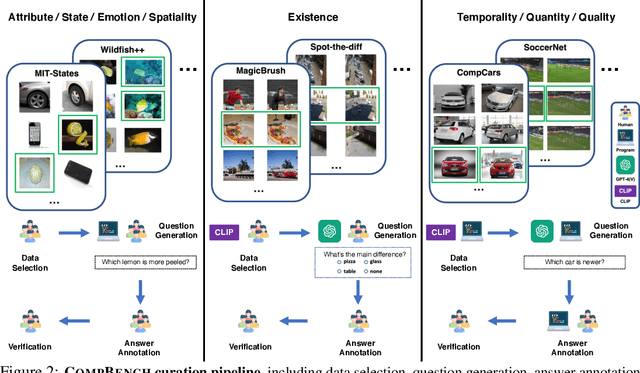
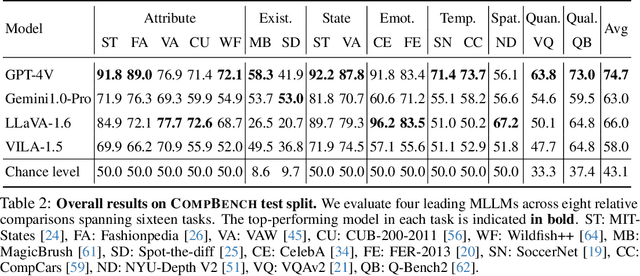
Abstract:The ability to compare objects, scenes, or situations is crucial for effective decision-making and problem-solving in everyday life. For instance, comparing the freshness of apples enables better choices during grocery shopping, while comparing sofa designs helps optimize the aesthetics of our living space. Despite its significance, the comparative capability is largely unexplored in artificial general intelligence (AGI). In this paper, we introduce CompBench, a benchmark designed to evaluate the comparative reasoning capability of multimodal large language models (MLLMs). CompBench mines and pairs images through visually oriented questions covering eight dimensions of relative comparison: visual attribute, existence, state, emotion, temporality, spatiality, quantity, and quality. We curate a collection of around 40K image pairs using metadata from diverse vision datasets and CLIP similarity scores. These image pairs span a broad array of visual domains, including animals, fashion, sports, and both outdoor and indoor scenes. The questions are carefully crafted to discern relative characteristics between two images and are labeled by human annotators for accuracy and relevance. We use CompBench to evaluate recent MLLMs, including GPT-4V(ision), Gemini-Pro, and LLaVA-1.6. Our results reveal notable shortcomings in their comparative abilities. We believe CompBench not only sheds light on these limitations but also establishes a solid foundation for future enhancements in the comparative capability of MLLMs.
Two-Stage Hybrid Supervision Framework for Fast, Low-resource, and Accurate Organ and Pan-cancer Segmentation in Abdomen CT
Sep 11, 2023



Abstract:Abdominal organ and tumour segmentation has many important clinical applications, such as organ quantification, surgical planning, and disease diagnosis. However, manual assessment is inherently subjective with considerable inter- and intra-expert variability. In the paper, we propose a hybrid supervised framework, StMt, that integrates self-training and mean teacher for the segmentation of abdominal organs and tumors using partially labeled and unlabeled data. We introduce a two-stage segmentation pipeline and whole-volume-based input strategy to maximize segmentation accuracy while meeting the requirements of inference time and GPU memory usage. Experiments on the validation set of FLARE2023 demonstrate that our method achieves excellent segmentation performance as well as fast and low-resource model inference. Our method achieved an average DSC score of 89.79\% and 45.55 \% for the organs and lesions on the validation set and the average running time and area under GPU memory-time cure are 11.25s and 9627.82MB, respectively.
TSI-Net: A Timing Sequence Image Segmentation Network for Intracranial Artery Segmentation in Digital Subtraction Angiography
Sep 07, 2023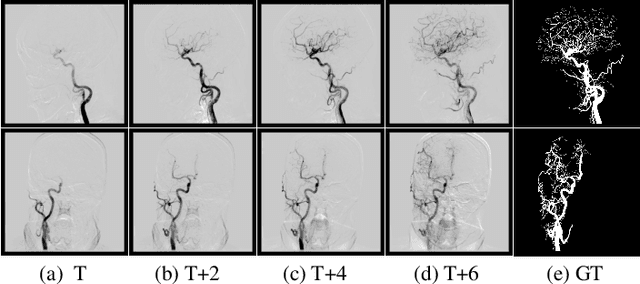
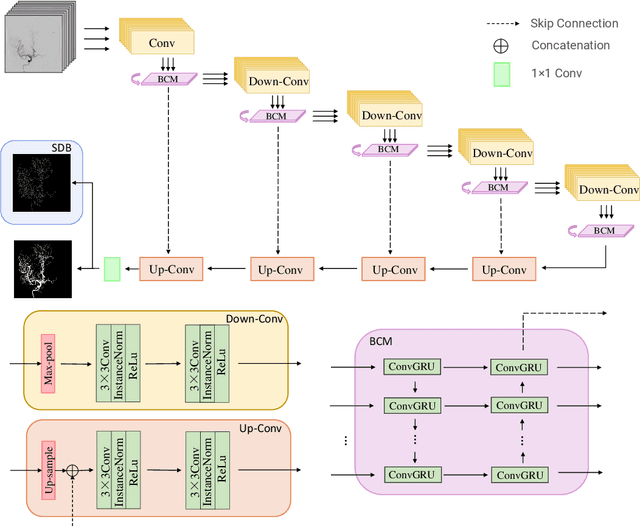

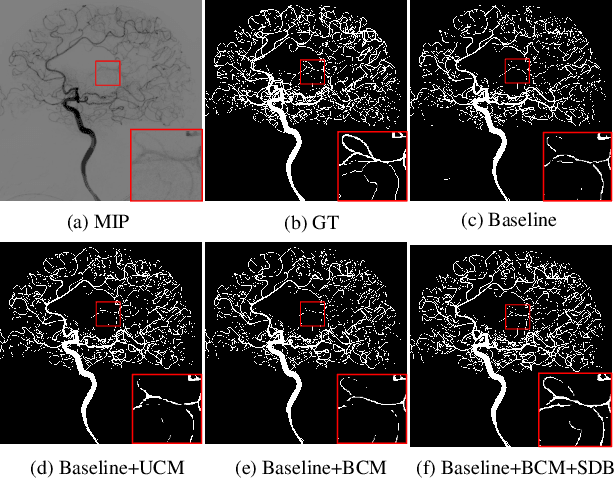
Abstract:Cerebrovascular disease is one of the major diseases facing the world today. Automatic segmentation of intracranial artery (IA) in digital subtraction angiography (DSA) sequences is an important step in the diagnosis of vascular related diseases and in guiding neurointerventional procedures. While, a single image can only show part of the IA within the contrast medium according to the imaging principle of DSA technology. Therefore, 2D DSA segmentation methods are unable to capture the complete IA information and treatment of cerebrovascular diseases. We propose A timing sequence image segmentation network with U-shape, called TSI-Net, which incorporates a bi-directional ConvGRU module (BCM) in the encoder. The network incorporates a bi-directional ConvGRU module (BCM) in the encoder, which can input variable-length DSA sequences, retain past and future information, segment them into 2D images. In addition, we introduce a sensitive detail branch (SDB) at the end for supervising fine vessels. Experimented on the DSA sequence dataset DIAS, the method performs significantly better than state-of-the-art networks in recent years. In particular, it achieves a Sen evaluation metric of 0.797, which is a 3% improvement compared to other methods.
DIAS: A Comprehensive Benchmark for DSA-sequence Intracranial Artery Segmentation
Jun 21, 2023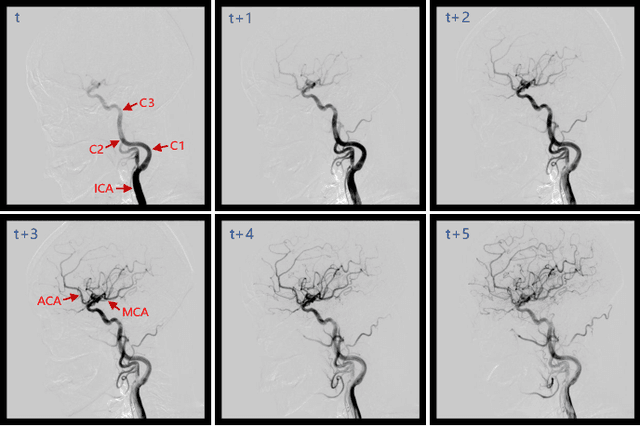
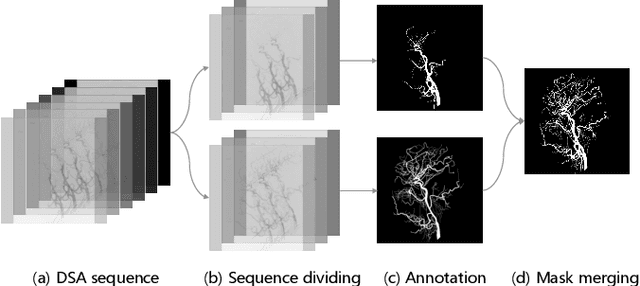
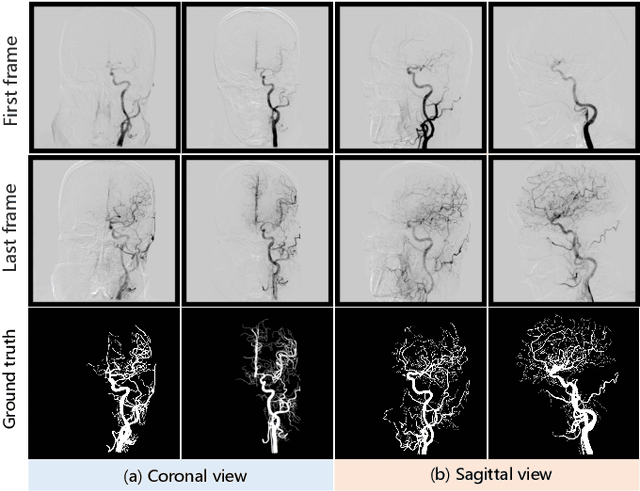
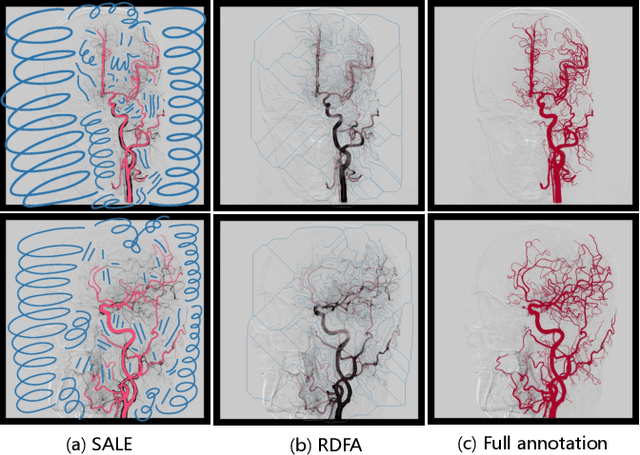
Abstract:Automatic segmentation of the intracranial artery (IA) in digital subtraction angiography (DSA) sequence is an essential step in diagnosing IA-related diseases and guiding neuro-interventional surgery. However, the lack of publicly available datasets has impeded research in this area. In this paper, we release DIAS, an IA segmentation dataset, consisting of 120 DSA sequences from intracranial interventional therapy. In addition to pixel-wise annotations, this dataset provides two types of scribble annotations for weakly supervised IA segmentation research. We present a comprehensive benchmark for evaluating the performance of this challenging dataset by utilizing fully-, weakly-, and semi-supervised learning approaches. Specifically, we propose a method that incorporates a dimensionality reduction module into a 2D/3D model to achieve vessel segmentation in DSA sequences. For weakly-supervised learning, we propose a scribble learning-based image segmentation framework, SSCR, which comprises scribble supervision and consistency regularization. Furthermore, we introduce a random patch-based self-training framework that utilizes unlabeled DSA sequences to improve segmentation performance. Our extensive experiments on the DIAS dataset demonstrate the effectiveness of these methods as potential baselines for future research and clinical applications.
Combining Hybrid Architecture and Pseudo-label for Semi-supervised Abdominal Organ Segmentation
Jul 23, 2022

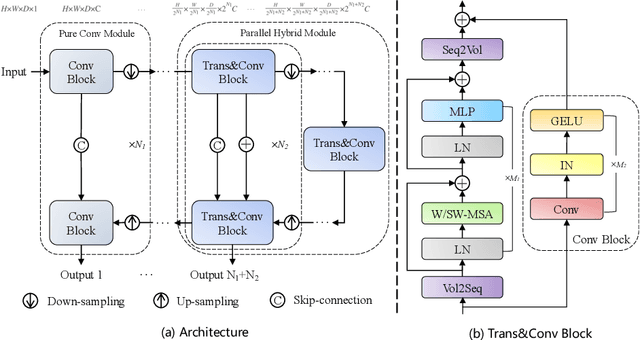
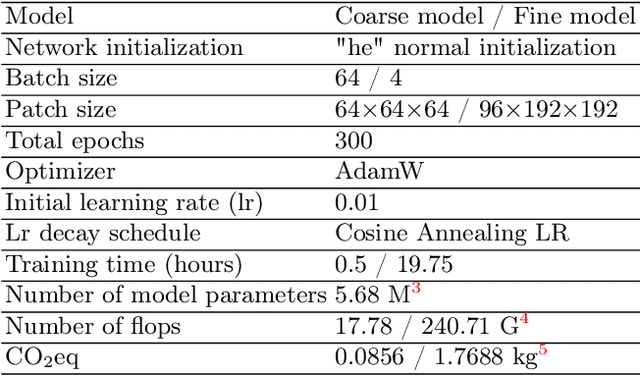
Abstract:Abdominal organ segmentation has many important clinical applications, such as organ quantification, surgical planning, and disease diagnosis. However, manually annotating organs from CT scans is time-consuming and labor-intensive. Semi-supervised learning has shown the potential to alleviate this challenge by learning from a large set of unlabeled images and limited labeled samples. In this work, we follow the self-training strategy and employ a hybrid architecture (PHTrans) with CNN and Transformer for both teacher and student models to generate precise pseudo-labels. Afterward, we introduce them with label data together into a two-stage segmentation framework with lightweight PHTrans for training to improve the performance and generalization ability of the model while remaining efficient. Experiments on the validation set of FLARE2022 demonstrate that our method achieves excellent segmentation performance as well as fast and low-resource model inference. The average DSC and HSD are 0.8956 and 0.9316, respectively. Under our development environments, the average inference time is 18.62 s, the average maximum GPU memory is 1995.04 MB, and the area under the GPU memory-time curve and the average area under the CPU utilization-time curve are 23196.84 and 319.67.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge