Tong Tian
Multi-wavelength ghost imaging: a review
Jun 16, 2025Abstract:Ghost imaging (GI) forms images from intensity-correlation data collected by a single-pixel detector, decoupling illumination and sensing. Since its quantum-photon origins, the technique has evolved through classical pseudothermal, computational and deep-learning variants to span an unprecedented spectral range--from extreme-ultraviolet (XUV) to terahertz (THz) waves and even matter waves. This review traces that evolution, highlighting how wavelength dictates modulators, detectors and propagation physics and, in turn, the attainable penetration depth, resolution and dose. We survey X-ray/XUV implementations that deliver low-damage microscopy, visible/near-IR systems that achieve video-rate lidar through fog and water, mid-IR platforms that extract molecular fingerprints in photon-starved conditions, and THz schemes that provide non-destructive inspection of concealed structures. These advances collectively position multi-wavelength GI as a versatile, low-dose alternative wherever conventional focal-plane arrays falter.
Image-Guided Autonomous Guidewire Navigation in Robot-Assisted Endovascular Interventions using Reinforcement Learning
Mar 09, 2024



Abstract:Autonomous robots in endovascular interventions possess the potential to navigate guidewires with safety and reliability, while reducing human error and shortening surgical time. However, current methods of guidewire navigation based on Reinforcement Learning (RL) depend on manual demonstration data or magnetic guidance. In this work, we propose an Image-guided Autonomous Guidewire Navigation (IAGN) method. Specifically, we introduce BDA-star, a path planning algorithm with boundary distance constraints, for the trajectory planning of guidewire navigation. We established an IAGN-RL environment where the observations are real-time guidewire feeding images highlighting the position of the guidewire tip and the planned path. We proposed a reward function based on the distances from both the guidewire tip to the planned path and the target to evaluate the agent's actions. Furthermore, in policy network, we employ a pre-trained convolutional neural network to extract features, mitigating stability issues and slow convergence rates associated with direct learning from raw pixels. Experiments conducted on the aortic simulation IAGN platform demonstrated that the proposed method, targeting the left subclavian artery and the brachiocephalic artery, achieved a 100% guidewire navigation success rate, along with reduced movement and retraction distances and trajectories tend to the center of the vessels.
UDCR: Unsupervised Aortic DSA/CTA Rigid Registration Using Deep Reinforcement Learning and Overlap Degree Calculation
Mar 09, 2024



Abstract:The rigid registration of aortic Digital Subtraction Angiography (DSA) and Computed Tomography Angiography (CTA) can provide 3D anatomical details of the vasculature for the interventional surgical treatment of conditions such as aortic dissection and aortic aneurysms, holding significant value for clinical research. However, the current methods for 2D/3D image registration are dependent on manual annotations or synthetic data, as well as the extraction of landmarks, which is not suitable for cross-modal registration of aortic DSA/CTA. In this paper, we propose an unsupervised method, UDCR, for aortic DSA/CTA rigid registration based on deep reinforcement learning. Leveraging the imaging principles and characteristics of DSA and CTA, we have constructed a cross-dimensional registration environment based on spatial transformations. Specifically, we propose an overlap degree calculation reward function that measures the intensity difference between the foreground and background, aimed at assessing the accuracy of registration between segmentation maps and DSA images. This method is highly flexible, allowing for the loading of pre-trained models to perform registration directly or to seek the optimal spatial transformation parameters through online learning. We manually annotated 61 pairs of aortic DSA/CTA for algorithm evaluation. The results indicate that the proposed UDCR achieved a Mean Absolute Error (MAE) of 2.85 mm in translation and 4.35{\deg} in rotation, showing significant potential for clinical applications.
Two-Stage Hybrid Supervision Framework for Fast, Low-resource, and Accurate Organ and Pan-cancer Segmentation in Abdomen CT
Sep 11, 2023



Abstract:Abdominal organ and tumour segmentation has many important clinical applications, such as organ quantification, surgical planning, and disease diagnosis. However, manual assessment is inherently subjective with considerable inter- and intra-expert variability. In the paper, we propose a hybrid supervised framework, StMt, that integrates self-training and mean teacher for the segmentation of abdominal organs and tumors using partially labeled and unlabeled data. We introduce a two-stage segmentation pipeline and whole-volume-based input strategy to maximize segmentation accuracy while meeting the requirements of inference time and GPU memory usage. Experiments on the validation set of FLARE2023 demonstrate that our method achieves excellent segmentation performance as well as fast and low-resource model inference. Our method achieved an average DSC score of 89.79\% and 45.55 \% for the organs and lesions on the validation set and the average running time and area under GPU memory-time cure are 11.25s and 9627.82MB, respectively.
DIAS: A Comprehensive Benchmark for DSA-sequence Intracranial Artery Segmentation
Jun 21, 2023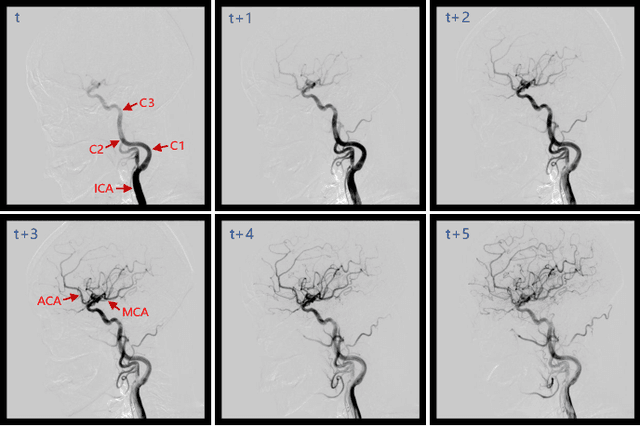
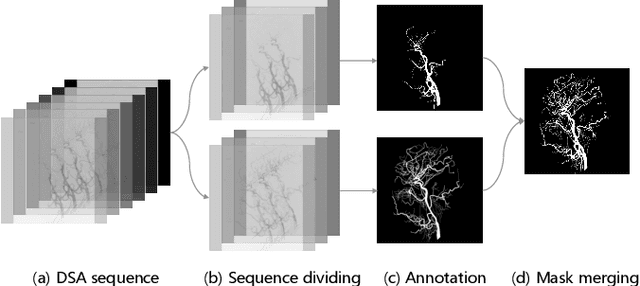
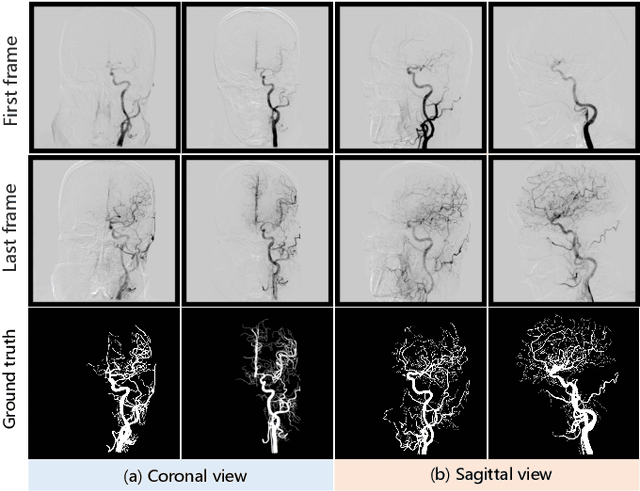
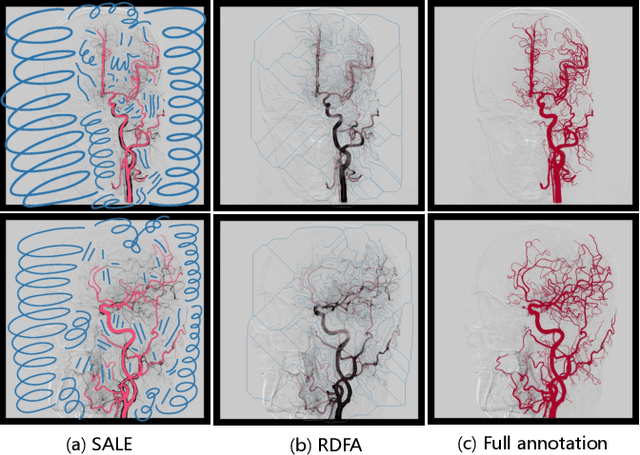
Abstract:Automatic segmentation of the intracranial artery (IA) in digital subtraction angiography (DSA) sequence is an essential step in diagnosing IA-related diseases and guiding neuro-interventional surgery. However, the lack of publicly available datasets has impeded research in this area. In this paper, we release DIAS, an IA segmentation dataset, consisting of 120 DSA sequences from intracranial interventional therapy. In addition to pixel-wise annotations, this dataset provides two types of scribble annotations for weakly supervised IA segmentation research. We present a comprehensive benchmark for evaluating the performance of this challenging dataset by utilizing fully-, weakly-, and semi-supervised learning approaches. Specifically, we propose a method that incorporates a dimensionality reduction module into a 2D/3D model to achieve vessel segmentation in DSA sequences. For weakly-supervised learning, we propose a scribble learning-based image segmentation framework, SSCR, which comprises scribble supervision and consistency regularization. Furthermore, we introduce a random patch-based self-training framework that utilizes unlabeled DSA sequences to improve segmentation performance. Our extensive experiments on the DIAS dataset demonstrate the effectiveness of these methods as potential baselines for future research and clinical applications.
PHTrans: Parallelly Aggregating Global and Local Representations for Medical Image Segmentation
Mar 13, 2022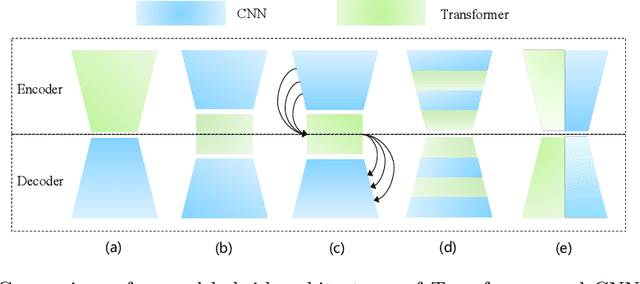
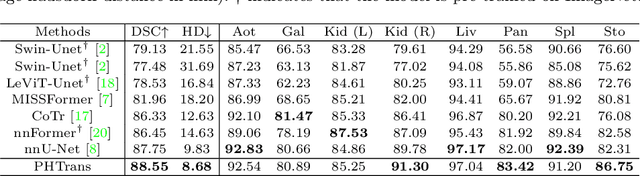
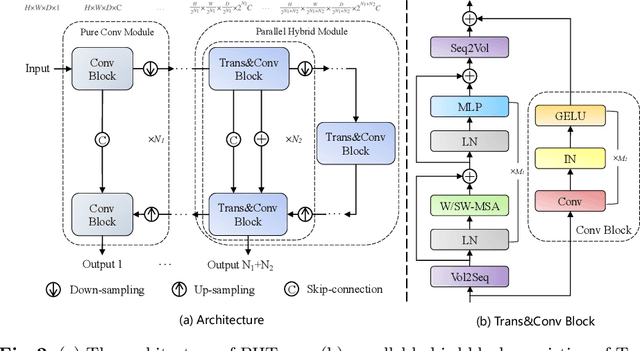
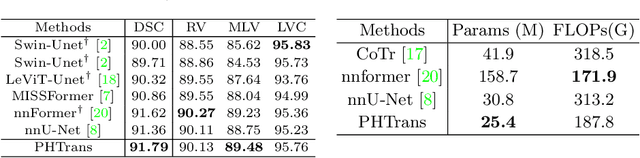
Abstract:The success of Transformer in computer vision has attracted increasing attention in the medical imaging community. Especially for medical image segmentation, many excellent hybrid architectures based on convolutional neural networks (CNNs) and Transformer have been presented and achieve impressive performance. However, most of these methods, which embed modular Transformer into CNNs, struggle to reach their full potential. In this paper, we propose a novel hybrid architecture for medical image segmentation called PHTrans, which parallelly hybridizes Transformer and CNN in main building blocks to produce hierarchical representations from global and local features and adaptively aggregate them, aiming to fully exploit their strengths to obtain better segmentation performance. Specifically, PHTrans follows the U-shaped encoder-decoder design and introduces the parallel hybird module in deep stages, where convolution blocks and the modified 3D Swin Transformer learn local features and global dependencies separately, then a sequence-to-volume operation unifies the dimensions of the outputs to achieve feature aggregation. Extensive experimental results on both Multi-Atlas Labeling Beyond the Cranial Vault and Automated Cardiac Diagnosis Challeng datasets corroborate its effectiveness, consistently outperforming state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge