Mohannad Elhamod
Data-to-Dashboard: Multi-Agent LLM Framework for Insightful Visualization in Enterprise Analytics
May 29, 2025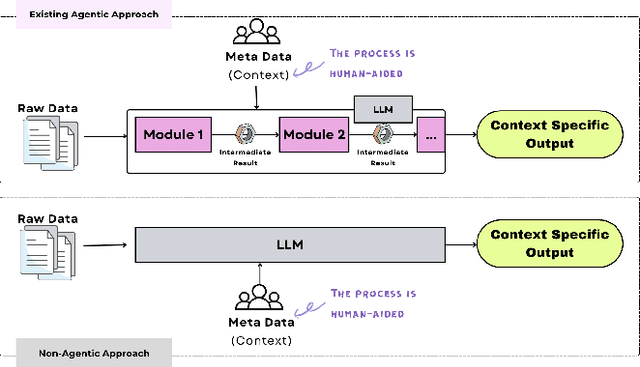
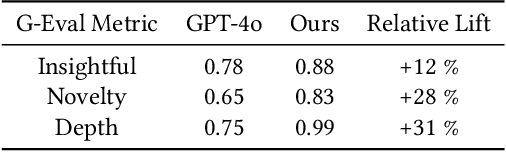
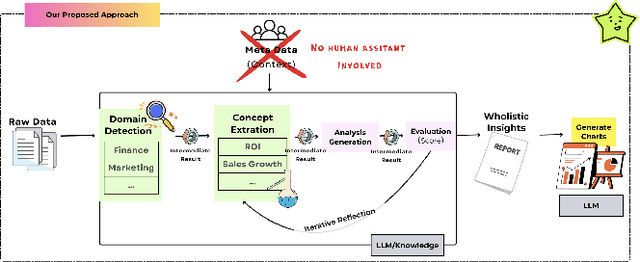
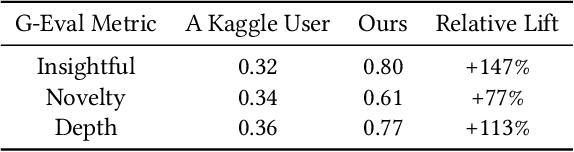
Abstract:The rapid advancement of LLMs has led to the creation of diverse agentic systems in data analysis, utilizing LLMs' capabilities to improve insight generation and visualization. In this paper, we present an agentic system that automates the data-to-dashboard pipeline through modular LLM agents capable of domain detection, concept extraction, multi-perspective analysis generation, and iterative self-reflection. Unlike existing chart QA systems, our framework simulates the analytical reasoning process of business analysts by retrieving domain-relevant knowledge and adapting to diverse datasets without relying on closed ontologies or question templates. We evaluate our system on three datasets across different domains. Benchmarked against GPT-4o with a single-prompt baseline, our approach shows improved insightfulness, domain relevance, and analytical depth, as measured by tailored evaluation metrics and qualitative human assessment. This work contributes a novel modular pipeline to bridge the path from raw data to visualization, and opens new opportunities for human-in-the-loop validation by domain experts in business analytics. All code can be found here: https://github.com/77luvC/D2D_Data2Dashboard
Neuro-Visualizer: An Auto-encoder-based Loss Landscape Visualization Method
Sep 26, 2023Abstract:In recent years, there has been a growing interest in visualizing the loss landscape of neural networks. Linear landscape visualization methods, such as principal component analysis, have become widely used as they intuitively help researchers study neural networks and their training process. However, these linear methods suffer from limitations and drawbacks due to their lack of flexibility and low fidelity at representing the high dimensional landscape. In this paper, we present a novel auto-encoder-based non-linear landscape visualization method called Neuro-Visualizer that addresses these shortcoming and provides useful insights about neural network loss landscapes. To demonstrate its potential, we run experiments on a variety of problems in two separate applications of knowledge-guided machine learning (KGML). Our findings show that Neuro-Visualizer outperforms other linear and non-linear baselines and helps corroborate, and sometime challenge, claims proposed by machine learning community. All code and data used in the experiments of this paper are available at an anonymous link https://anonymous.4open.science/r/NeuroVisualizer-FDD6
Discovering Novel Biological Traits From Images Using Phylogeny-Guided Neural Networks
Jun 05, 2023



Abstract:Discovering evolutionary traits that are heritable across species on the tree of life (also referred to as a phylogenetic tree) is of great interest to biologists to understand how organisms diversify and evolve. However, the measurement of traits is often a subjective and labor-intensive process, making trait discovery a highly label-scarce problem. We present a novel approach for discovering evolutionary traits directly from images without relying on trait labels. Our proposed approach, Phylo-NN, encodes the image of an organism into a sequence of quantized feature vectors -- or codes -- where different segments of the sequence capture evolutionary signals at varying ancestry levels in the phylogeny. We demonstrate the effectiveness of our approach in producing biologically meaningful results in a number of downstream tasks including species image generation and species-to-species image translation, using fish species as a target example.
Learning Neural Networks with Competing Physics Objectives: An Application in Quantum Mechanics
Jul 02, 2020
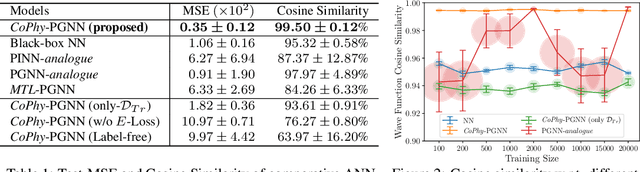


Abstract:Physics-guided Machine Learning (PGML) is an emerging field of research in machine learning (ML) that aims to harness the power of ML advances without ignoring the rich knowledge of physics underlying scientific phenomena. One of the promising directions in PGML is to modify the objective function of neural networks by adding physics-guided (PG) loss functions that measure the violation of physics objectives in the ANN outputs. Existing PGML approaches generally focus on incorporating a single physics objective as a PG loss, using constant trade-off parameters. However, in the presence of multiple physics objectives with competing non-convex PG loss terms, there is a need to adaptively tune the importance of competing PG loss terms during the process of neural network training. We present a novel approach to handle competing PG loss terms in the illustrative application of quantum mechanics, where the two competing physics objectives are minimizing the energy while satisfying the Schrodinger equation. We conducted a systematic evaluation of the effects of PG loss on the generalization ability of neural networks in comparison with several baseline methods in PGML. All the code and data used in this work is available at https://github.com/jayroxis/Cophy-PGNN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge