Charles Stewart
Fine-Tuning is Fine, if Calibrated
Sep 24, 2024



Abstract:Fine-tuning is arguably the most straightforward way to tailor a pre-trained model (e.g., a foundation model) to downstream applications, but it also comes with the risk of losing valuable knowledge the model had learned in pre-training. For example, fine-tuning a pre-trained classifier capable of recognizing a large number of classes to master a subset of classes at hand is shown to drastically degrade the model's accuracy in the other classes it had previously learned. As such, it is hard to further use the fine-tuned model when it encounters classes beyond the fine-tuning data. In this paper, we systematically dissect the issue, aiming to answer the fundamental question, ''What has been damaged in the fine-tuned model?'' To our surprise, we find that the fine-tuned model neither forgets the relationship among the other classes nor degrades the features to recognize these classes. Instead, the fine-tuned model often produces more discriminative features for these other classes, even if they were missing during fine-tuning! {What really hurts the accuracy is the discrepant logit scales between the fine-tuning classes and the other classes}, implying that a simple post-processing calibration would bring back the pre-trained model's capability and at the same time unveil the feature improvement over all classes. We conduct an extensive empirical study to demonstrate the robustness of our findings and provide preliminary explanations underlying them, suggesting new directions for future theoretical analysis. Our code is available at https://github.com/OSU-MLB/Fine-Tuning-Is-Fine-If-Calibrated.
VLM4Bio: A Benchmark Dataset to Evaluate Pretrained Vision-Language Models for Trait Discovery from Biological Images
Aug 28, 2024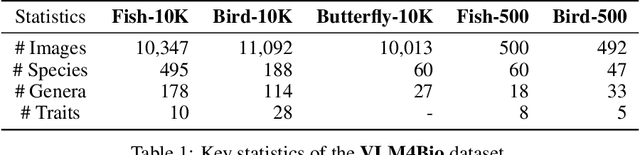
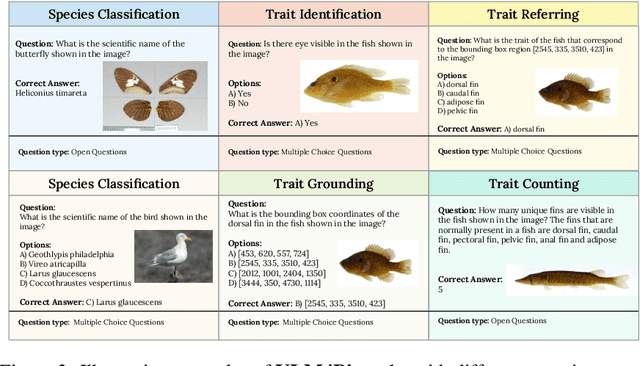

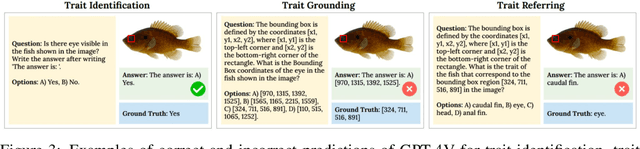
Abstract:Images are increasingly becoming the currency for documenting biodiversity on the planet, providing novel opportunities for accelerating scientific discoveries in the field of organismal biology, especially with the advent of large vision-language models (VLMs). We ask if pre-trained VLMs can aid scientists in answering a range of biologically relevant questions without any additional fine-tuning. In this paper, we evaluate the effectiveness of 12 state-of-the-art (SOTA) VLMs in the field of organismal biology using a novel dataset, VLM4Bio, consisting of 469K question-answer pairs involving 30K images from three groups of organisms: fishes, birds, and butterflies, covering five biologically relevant tasks. We also explore the effects of applying prompting techniques and tests for reasoning hallucination on the performance of VLMs, shedding new light on the capabilities of current SOTA VLMs in answering biologically relevant questions using images. The code and datasets for running all the analyses reported in this paper can be found at https://github.com/sammarfy/VLM4Bio.
Hierarchical Conditioning of Diffusion Models Using Tree-of-Life for Studying Species Evolution
Jul 31, 2024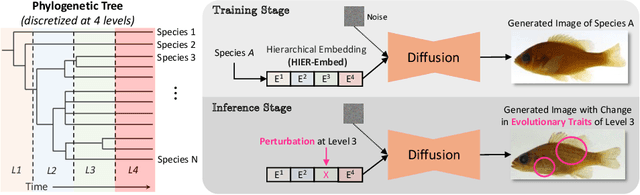
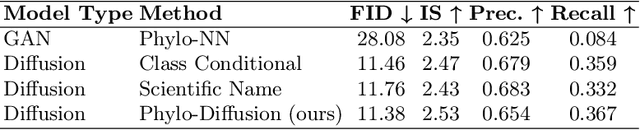
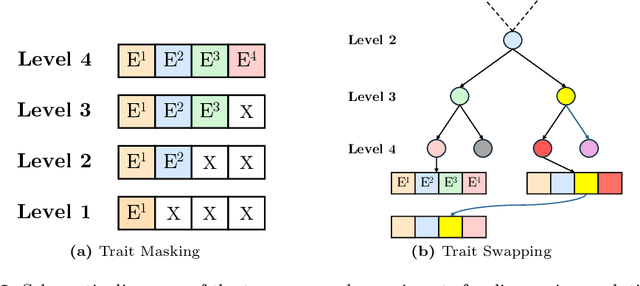

Abstract:A central problem in biology is to understand how organisms evolve and adapt to their environment by acquiring variations in the observable characteristics or traits of species across the tree of life. With the growing availability of large-scale image repositories in biology and recent advances in generative modeling, there is an opportunity to accelerate the discovery of evolutionary traits automatically from images. Toward this goal, we introduce Phylo-Diffusion, a novel framework for conditioning diffusion models with phylogenetic knowledge represented in the form of HIERarchical Embeddings (HIER-Embeds). We also propose two new experiments for perturbing the embedding space of Phylo-Diffusion: trait masking and trait swapping, inspired by counterpart experiments of gene knockout and gene editing/swapping. Our work represents a novel methodological advance in generative modeling to structure the embedding space of diffusion models using tree-based knowledge. Our work also opens a new chapter of research in evolutionary biology by using generative models to visualize evolutionary changes directly from images. We empirically demonstrate the usefulness of Phylo-Diffusion in capturing meaningful trait variations for fishes and birds, revealing novel insights about the biological mechanisms of their evolution.
Bringing Back the Context: Camera Trap Species Identification as Link Prediction on Multimodal Knowledge Graphs
Jan 08, 2024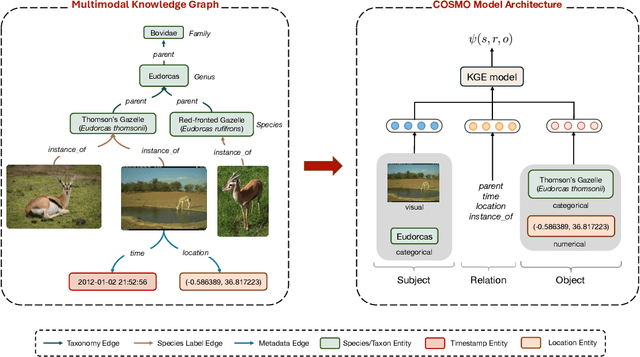
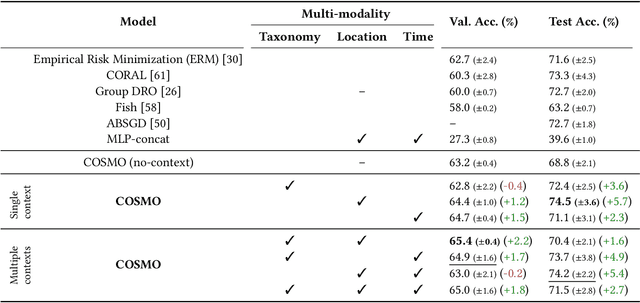
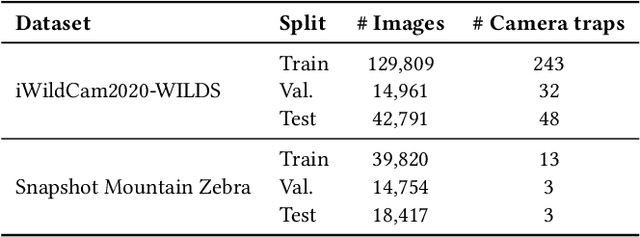
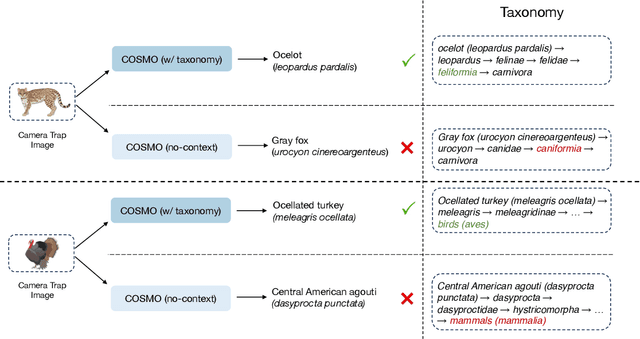
Abstract:Camera traps are valuable tools in animal ecology for biodiversity monitoring and conservation. However, challenges like poor generalization to deployment at new unseen locations limit their practical application. Images are naturally associated with heterogeneous forms of context possibly in different modalities. In this work, we leverage the structured context associated with the camera trap images to improve out-of-distribution generalization for the task of species identification in camera traps. For example, a photo of a wild animal may be associated with information about where and when it was taken, as well as structured biology knowledge about the animal species. While typically overlooked by existing work, bringing back such context offers several potential benefits for better image understanding, such as addressing data scarcity and enhancing generalization. However, effectively integrating such heterogeneous context into the visual domain is a challenging problem. To address this, we propose a novel framework that reformulates species classification as link prediction in a multimodal knowledge graph (KG). This framework seamlessly integrates various forms of multimodal context for visual recognition. We apply this framework for out-of-distribution species classification on the iWildCam2020-WILDS and Snapshot Mountain Zebra datasets and achieve competitive performance with state-of-the-art approaches. Furthermore, our framework successfully incorporates biological taxonomy for improved generalization and enhances sample efficiency for recognizing under-represented species.
BioCLIP: A Vision Foundation Model for the Tree of Life
Dec 04, 2023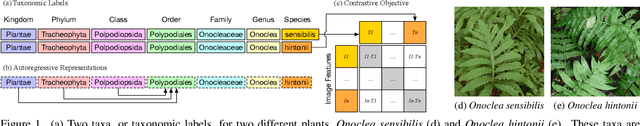
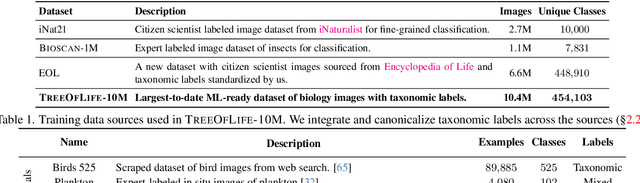
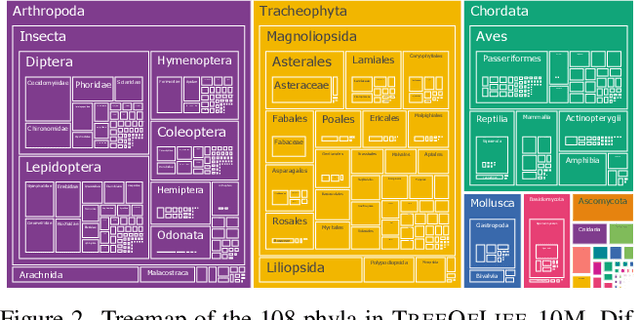
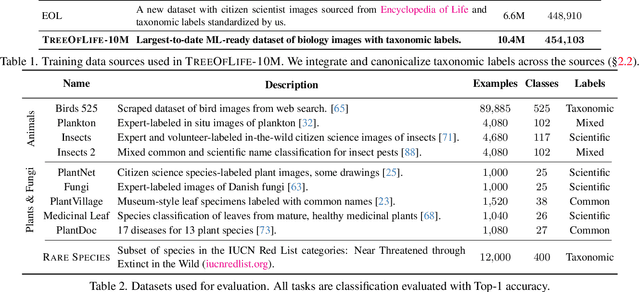
Abstract:Images of the natural world, collected by a variety of cameras, from drones to individual phones, are increasingly abundant sources of biological information. There is an explosion of computational methods and tools, particularly computer vision, for extracting biologically relevant information from images for science and conservation. Yet most of these are bespoke approaches designed for a specific task and are not easily adaptable or extendable to new questions, contexts, and datasets. A vision model for general organismal biology questions on images is of timely need. To approach this, we curate and release TreeOfLife-10M, the largest and most diverse ML-ready dataset of biology images. We then develop BioCLIP, a foundation model for the tree of life, leveraging the unique properties of biology captured by TreeOfLife-10M, namely the abundance and variety of images of plants, animals, and fungi, together with the availability of rich structured biological knowledge. We rigorously benchmark our approach on diverse fine-grained biology classification tasks, and find that BioCLIP consistently and substantially outperforms existing baselines (by 17% to 20% absolute). Intrinsic evaluation reveals that BioCLIP has learned a hierarchical representation conforming to the tree of life, shedding light on its strong generalizability. Our code, models and data will be made available at https://github.com/Imageomics/bioclip.
A Simple Interpretable Transformer for Fine-Grained Image Classification and Analysis
Nov 07, 2023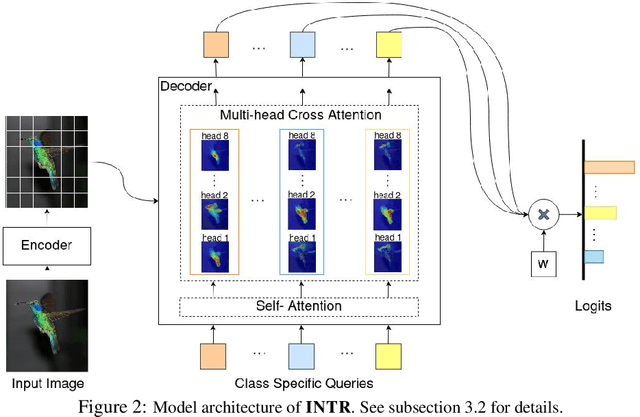
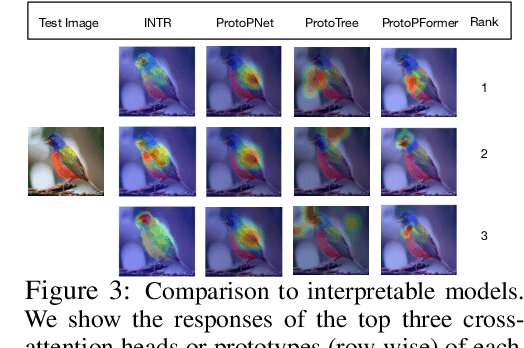
Abstract:We present a novel usage of Transformers to make image classification interpretable. Unlike mainstream classifiers that wait until the last fully-connected layer to incorporate class information to make predictions, we investigate a proactive approach, asking each class to search for itself in an image. We realize this idea via a Transformer encoder-decoder inspired by DEtection TRansformer (DETR). We learn ``class-specific'' queries (one for each class) as input to the decoder, enabling each class to localize its patterns in an image via cross-attention. We name our approach INterpretable TRansformer (INTR), which is fairly easy to implement and exhibits several compelling properties. We show that INTR intrinsically encourages each class to attend distinctively; the cross-attention weights thus provide a faithful interpretation of the prediction. Interestingly, via ``multi-head'' cross-attention, INTR could identify different ``attributes'' of a class, making it particularly suitable for fine-grained classification and analysis, which we demonstrate on eight datasets. Our code and pre-trained model are publicly accessible at https://github.com/Imageomics/INTR.
Holistic Transfer: Towards Non-Disruptive Fine-Tuning with Partial Target Data
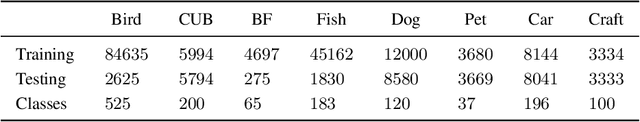
Nov 02, 2023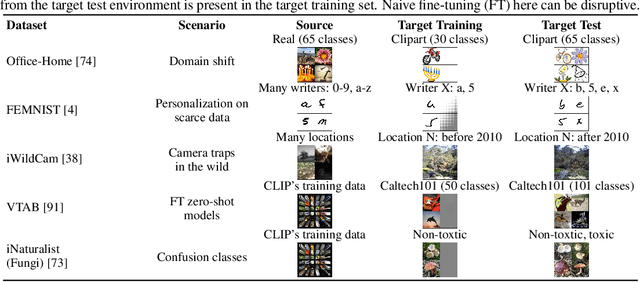
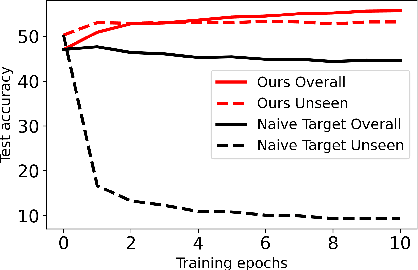
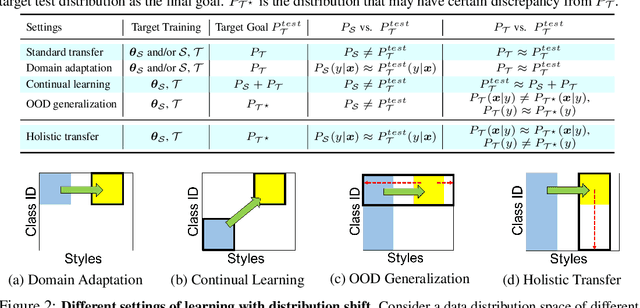
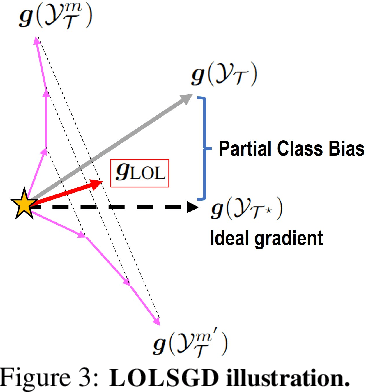
Abstract:We propose a learning problem involving adapting a pre-trained source model to the target domain for classifying all classes that appeared in the source data, using target data that covers only a partial label space. This problem is practical, as it is unrealistic for the target end-users to collect data for all classes prior to adaptation. However, it has received limited attention in the literature. To shed light on this issue, we construct benchmark datasets and conduct extensive experiments to uncover the inherent challenges. We found a dilemma -- on the one hand, adapting to the new target domain is important to claim better performance; on the other hand, we observe that preserving the classification accuracy of classes missing in the target adaptation data is highly challenging, let alone improving them. To tackle this, we identify two key directions: 1) disentangling domain gradients from classification gradients, and 2) preserving class relationships. We present several effective solutions that maintain the accuracy of the missing classes and enhance the overall performance, establishing solid baselines for holistic transfer of pre-trained models with partial target data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge