Mufei Li
Graph-KV: Breaking Sequence via Injecting Structural Biases into Large Language Models
Jun 09, 2025Abstract:Modern large language models (LLMs) are inherently auto-regressive, requiring input to be serialized into flat sequences regardless of their structural dependencies. This serialization hinders the model's ability to leverage structural inductive biases, especially in tasks such as retrieval-augmented generation (RAG) and reasoning on data with native graph structures, where inter-segment dependencies are crucial. We introduce Graph-KV with the potential to overcome this limitation. Graph-KV leverages the KV-cache of text segments as condensed representations and governs their interaction through structural inductive biases. In this framework, 'target' segments selectively attend only to the KV-caches of their designated 'source' segments, rather than all preceding segments in a serialized sequence. This approach induces a graph-structured block mask, sparsifying attention and enabling a message-passing-like step within the LLM. Furthermore, strategically allocated positional encodings for source and target segments reduce positional bias and context window consumption. We evaluate Graph-KV across three scenarios: (1) seven RAG benchmarks spanning direct inference, multi-hop reasoning, and long-document understanding; (2) Arxiv-QA, a novel academic paper QA task with full-text scientific papers structured as citation ego-graphs; and (3) paper topic classification within a citation network. By effectively reducing positional bias and harnessing structural inductive biases, Graph-KV substantially outperforms baselines, including standard costly sequential encoding, across various settings. Code and the Graph-KV data are publicly available.
Underestimated Privacy Risks for Minority Populations in Large Language Model Unlearning
Dec 11, 2024Abstract:Large Language Models are trained on extensive datasets that often contain sensitive, human-generated information, raising significant concerns about privacy breaches. While certified unlearning approaches offer strong privacy guarantees, they rely on restrictive model assumptions that are not applicable to LLMs. As a result, various unlearning heuristics have been proposed, with the associated privacy risks assessed only empirically. The standard evaluation pipelines typically randomly select data for removal from the training set, apply unlearning techniques, and use membership inference attacks to compare the unlearned models against models retrained without the to-be-unlearned data. However, since every data point is subject to the right to be forgotten, unlearning should be considered in the worst-case scenario from the privacy perspective. Prior work shows that data outliers may exhibit higher memorization effects. Intuitively, they are harder to be unlearn and thus the privacy risk of unlearning them is underestimated in the current evaluation. In this paper, we leverage minority data to identify such a critical flaw in previously widely adopted evaluations. We substantiate this claim through carefully designed experiments, including unlearning canaries related to minority groups, inspired by privacy auditing literature. Using personally identifiable information as a representative minority identifier, we demonstrate that minority groups experience at least 20% more privacy leakage in most cases across six unlearning approaches, three MIAs, three benchmark datasets, and two LLMs of different scales. Given that the right to be forgotten should be upheld for every individual, we advocate for a more rigorous evaluation of LLM unlearning methods. Our minority-aware evaluation framework represents an initial step toward ensuring more equitable assessments of LLM unlearning efficacy.
LayerDAG: A Layerwise Autoregressive Diffusion Model for Directed Acyclic Graph Generation
Nov 04, 2024



Abstract:Directed acyclic graphs (DAGs) serve as crucial data representations in domains such as hardware synthesis and compiler/program optimization for computing systems. DAG generative models facilitate the creation of synthetic DAGs, which can be used for benchmarking computing systems while preserving intellectual property. However, generating realistic DAGs is challenging due to their inherent directional and logical dependencies. This paper introduces LayerDAG, an autoregressive diffusion model, to address these challenges. LayerDAG decouples the strong node dependencies into manageable units that can be processed sequentially. By interpreting the partial order of nodes as a sequence of bipartite graphs, LayerDAG leverages autoregressive generation to model directional dependencies and employs diffusion models to capture logical dependencies within each bipartite graph. Comparative analyses demonstrate that LayerDAG outperforms existing DAG generative models in both expressiveness and generalization, particularly for generating large-scale DAGs with up to 400 nodes-a critical scenario for system benchmarking. Extensive experiments on both synthetic and real-world flow graphs from various computing platforms show that LayerDAG generates valid DAGs with superior statistical properties and benchmarking performance. The synthetic DAGs generated by LayerDAG enhance the training of ML-based surrogate models, resulting in improved accuracy in predicting performance metrics of real-world DAGs across diverse computing platforms.
Simple is Effective: The Roles of Graphs and Large Language Models in Knowledge-Graph-Based Retrieval-Augmented Generation
Oct 28, 2024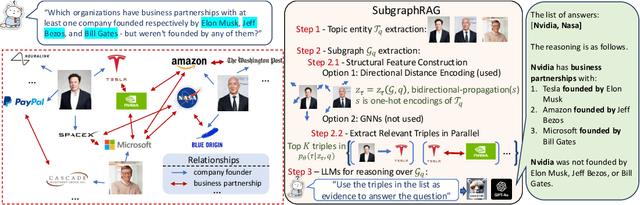
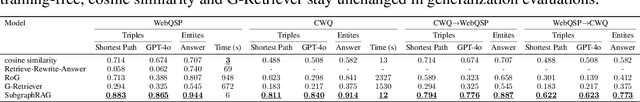
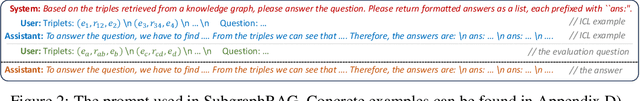
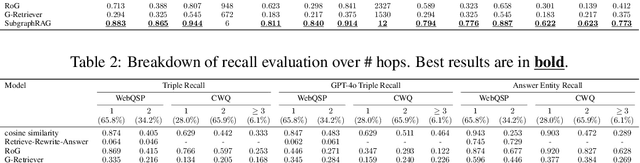
Abstract:Large Language Models (LLMs) demonstrate strong reasoning abilities but face limitations such as hallucinations and outdated knowledge. Knowledge Graph (KG)-based Retrieval-Augmented Generation (RAG) addresses these issues by grounding LLM outputs in structured external knowledge from KGs. However, current KG-based RAG frameworks still struggle to optimize the trade-off between retrieval effectiveness and efficiency in identifying a suitable amount of relevant graph information for the LLM to digest. We introduce SubgraphRAG, extending the KG-based RAG framework that retrieves subgraphs and leverages LLMs for reasoning and answer prediction. Our approach innovatively integrates a lightweight multilayer perceptron with a parallel triple-scoring mechanism for efficient and flexible subgraph retrieval while encoding directional structural distances to enhance retrieval effectiveness. The size of retrieved subgraphs can be flexibly adjusted to match the query's need and the downstream LLM's capabilities. This design strikes a balance between model complexity and reasoning power, enabling scalable and generalizable retrieval processes. Notably, based on our retrieved subgraphs, smaller LLMs like Llama3.1-8B-Instruct deliver competitive results with explainable reasoning, while larger models like GPT-4o achieve state-of-the-art accuracy compared with previous baselines -- all without fine-tuning. Extensive evaluations on the WebQSP and CWQ benchmarks highlight SubgraphRAG's strengths in efficiency, accuracy, and reliability by reducing hallucinations and improving response grounding.
KGExplainer: Towards Exploring Connected Subgraph Explanations for Knowledge Graph Completion
Apr 05, 2024


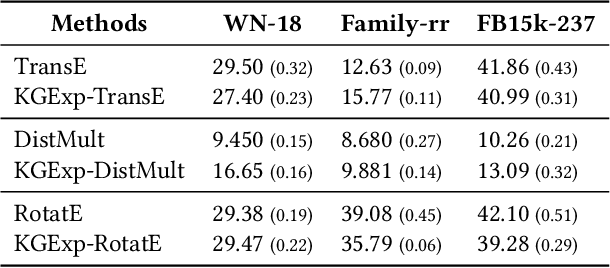
Abstract:Knowledge graph completion (KGC) aims to alleviate the inherent incompleteness of knowledge graphs (KGs), which is a critical task for various applications, such as recommendations on the web. Although knowledge graph embedding (KGE) models have demonstrated superior predictive performance on KGC tasks, these models infer missing links in a black-box manner that lacks transparency and accountability, preventing researchers from developing accountable models. Existing KGE-based explanation methods focus on exploring key paths or isolated edges as explanations, which is information-less to reason target prediction. Additionally, the missing ground truth leads to these explanation methods being ineffective in quantitatively evaluating explored explanations. To overcome these limitations, we propose KGExplainer, a model-agnostic method that identifies connected subgraph explanations and distills an evaluator to assess them quantitatively. KGExplainer employs a perturbation-based greedy search algorithm to find key connected subgraphs as explanations within the local structure of target predictions. To evaluate the quality of the explored explanations, KGExplainer distills an evaluator from the target KGE model. By forwarding the explanations to the evaluator, our method can examine the fidelity of them. Extensive experiments on benchmark datasets demonstrate that KGExplainer yields promising improvement and achieves an optimal ratio of 83.3% in human evaluation.
GraphMaker: Can Diffusion Models Generate Large Attributed Graphs?
Oct 20, 2023



Abstract:Large-scale graphs with node attributes are fundamental in real-world scenarios, such as social and financial networks. The generation of synthetic graphs that emulate real-world ones is pivotal in graph machine learning, aiding network evolution understanding and data utility preservation when original data cannot be shared. Traditional models for graph generation suffer from limited model capacity. Recent developments in diffusion models have shown promise in merely graph structure generation or the generation of small molecular graphs with attributes. However, their applicability to large attributed graphs remains unaddressed due to challenges in capturing intricate patterns and scalability. This paper introduces GraphMaker, a novel diffusion model tailored for generating large attributed graphs. We study the diffusion models that either couple or decouple graph structure and node attribute generation to address their complex correlation. We also employ node-level conditioning and adopt a minibatch strategy for scalability. We further propose a new evaluation pipeline using models trained on generated synthetic graphs and tested on original graphs to evaluate the quality of synthetic data. Empirical evaluations on real-world datasets showcase GraphMaker's superiority in generating realistic and diverse large-attributed graphs beneficial for downstream tasks.
Benchmarking Accuracy and Generalizability of Four Graph Neural Networks Using Large In Vitro ADME Datasets from Different Chemical Spaces
Nov 27, 2021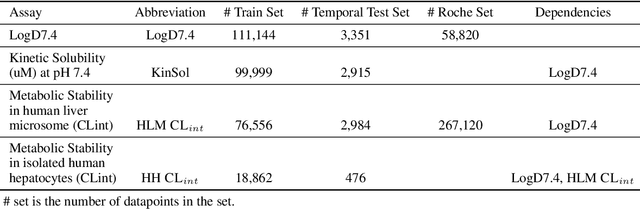
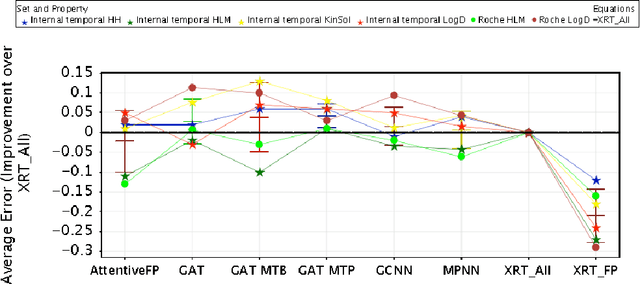
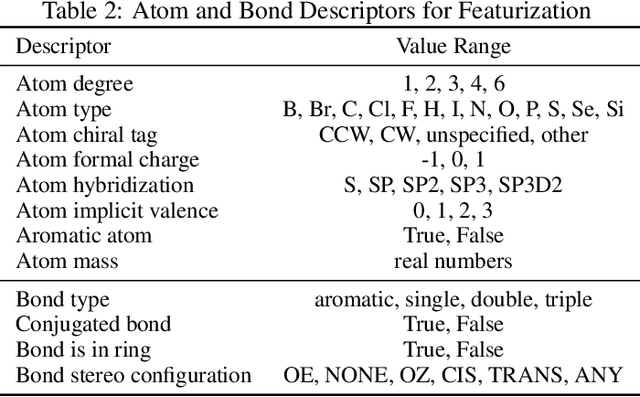
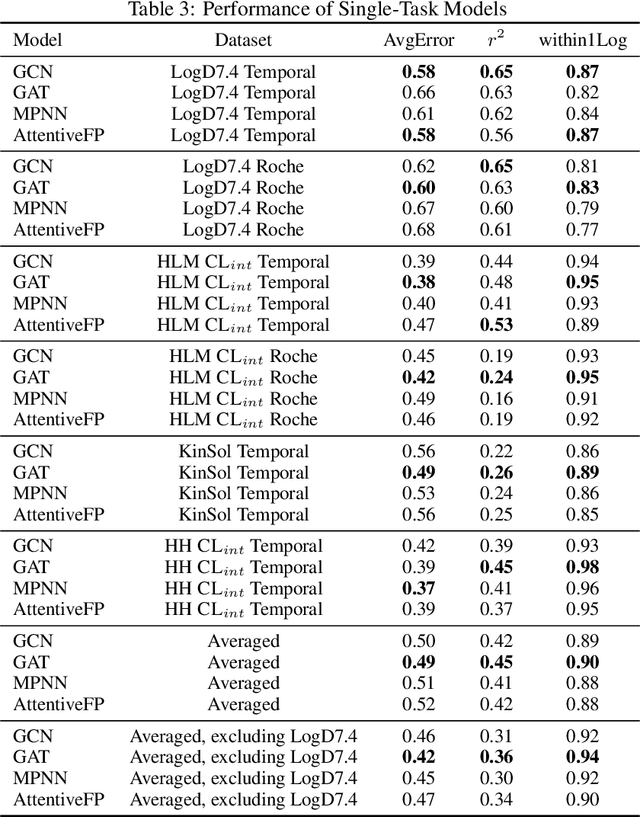
Abstract:In this work, we benchmark a variety of single- and multi-task graph neural network (GNN) models against lower-bar and higher-bar traditional machine learning approaches employing human engineered molecular features. We consider four GNN variants -- Graph Convolutional Network (GCN), Graph Attention Network (GAT), Message Passing Neural Network (MPNN), and Attentive Fingerprint (AttentiveFP). So far deep learning models have been primarily benchmarked using lower-bar traditional models solely based on fingerprints, while more realistic benchmarks employing fingerprints, whole-molecule descriptors and predictions from other related endpoints (e.g., LogD7.4) appear to be scarce for industrial ADME datasets. In addition to time-split test sets based on Genentech data, this study benefits from the availability of measurements from an external chemical space (Roche data). We identify GAT as a promising approach to implementing deep learning models. While all GNN models significantly outperform lower-bar benchmark traditional models solely based on fingerprints, only GATs seem to offer a small but consistent improvement over higher-bar benchmark traditional models. Finally, the accuracy of in vitro assays from different laboratories predicting the same experimental endpoints appears to be comparable with the accuracy of GAT single-task models, suggesting that most of the observed error from the models is a function of the experimental error propagation.
DGL-LifeSci: An Open-Source Toolkit for Deep Learning on Graphs in Life Science
Jun 27, 2021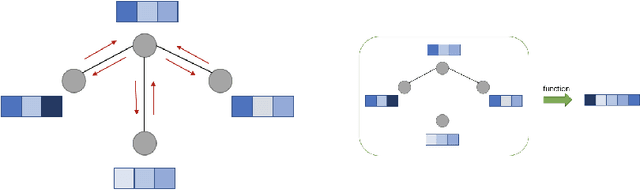
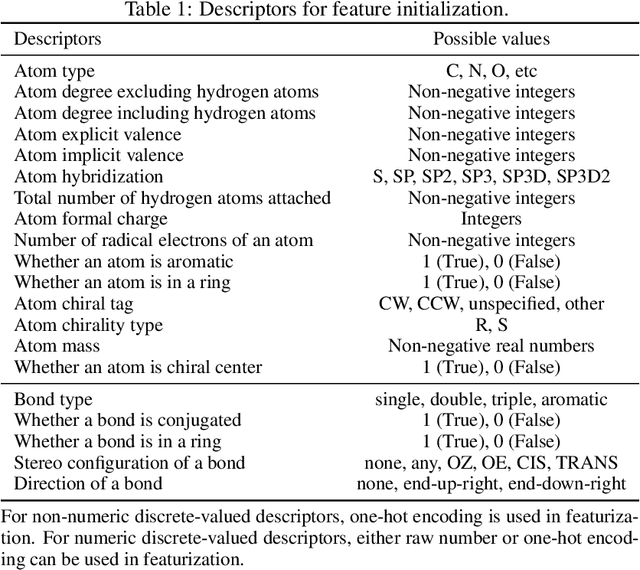
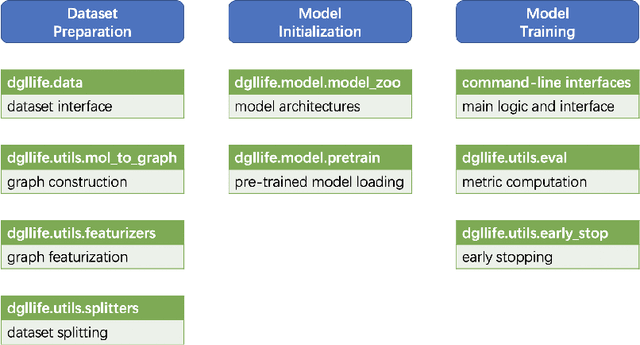

Abstract:Graph neural networks (GNNs) constitute a class of deep learning methods for graph data. They have wide applications in chemistry and biology, such as molecular property prediction, reaction prediction and drug-target interaction prediction. Despite the interest, GNN-based modeling is challenging as it requires graph data pre-processing and modeling in addition to programming and deep learning. Here we present DGL-LifeSci, an open-source package for deep learning on graphs in life science. DGL-LifeSci is a python toolkit based on RDKit, PyTorch and Deep Graph Library (DGL). DGL-LifeSci allows GNN-based modeling on custom datasets for molecular property prediction, reaction prediction and molecule generation. With its command-line interfaces, users can perform modeling without any background in programming and deep learning. We test the command-line interfaces using standard benchmarks MoleculeNet, USPTO, and ZINC. Compared with previous implementations, DGL-LifeSci achieves a speed up by up to 6x. For modeling flexibility, DGL-LifeSci provides well-optimized modules for various stages of the modeling pipeline. In addition, DGL-LifeSci provides pre-trained models for reproducing the test experiment results and applying models without training. The code is distributed under an Apache-2.0 License and is freely accessible at https://github.com/awslabs/dgl-lifesci.
Deep Graph Library: Towards Efficient and Scalable Deep Learning on Graphs
Sep 03, 2019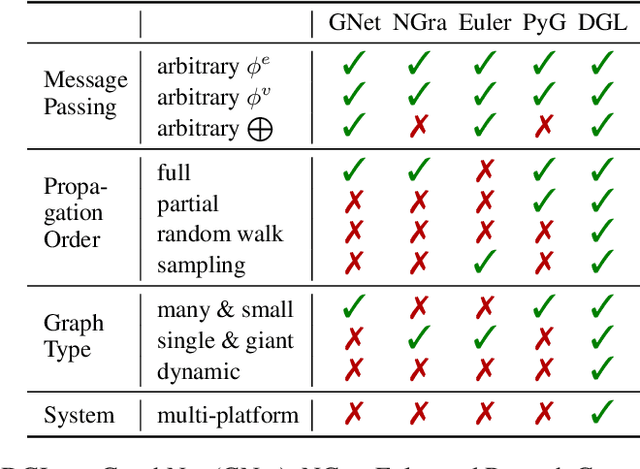
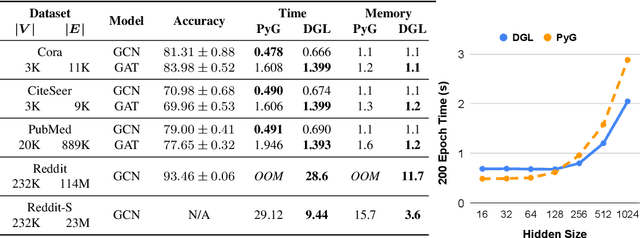
Abstract:Accelerating research in the emerging field of deep graph learning requires new tools. Such systems should support graph as the core abstraction and take care to maintain both forward (i.e. supporting new research ideas) and backward (i.e. integration with existing components) compatibility. In this paper, we present Deep Graph Library (DGL). DGL enables arbitrary message handling and mutation operators, flexible propagation rules, and is framework agnostic so as to leverage high-performance tensor, autograd operations, and other feature extraction modules already available in existing frameworks. DGL carefully handles the sparse and irregular graph structure, deals with graphs big and small which may change dynamically, fuses operations, and performs auto-batching, all to take advantages of modern hardware. DGL has been tested on a variety of models, including but not limited to the popular Graph Neural Networks (GNN) and its variants, with promising speed, memory footprint and scalability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge