Xiaoqin Pan
KGExplainer: Towards Exploring Connected Subgraph Explanations for Knowledge Graph Completion
Apr 05, 2024


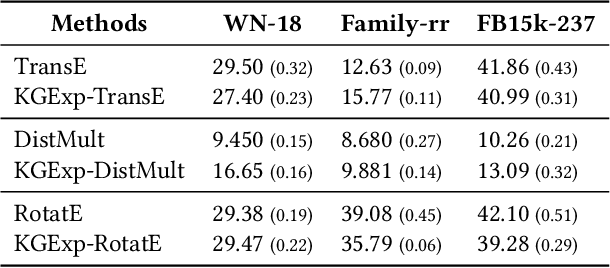
Abstract:Knowledge graph completion (KGC) aims to alleviate the inherent incompleteness of knowledge graphs (KGs), which is a critical task for various applications, such as recommendations on the web. Although knowledge graph embedding (KGE) models have demonstrated superior predictive performance on KGC tasks, these models infer missing links in a black-box manner that lacks transparency and accountability, preventing researchers from developing accountable models. Existing KGE-based explanation methods focus on exploring key paths or isolated edges as explanations, which is information-less to reason target prediction. Additionally, the missing ground truth leads to these explanation methods being ineffective in quantitatively evaluating explored explanations. To overcome these limitations, we propose KGExplainer, a model-agnostic method that identifies connected subgraph explanations and distills an evaluator to assess them quantitatively. KGExplainer employs a perturbation-based greedy search algorithm to find key connected subgraphs as explanations within the local structure of target predictions. To evaluate the quality of the explored explanations, KGExplainer distills an evaluator from the target KGE model. By forwarding the explanations to the evaluator, our method can examine the fidelity of them. Extensive experiments on benchmark datasets demonstrate that KGExplainer yields promising improvement and achieves an optimal ratio of 83.3% in human evaluation.
Deep learning for drug repurposing: methods, databases, and applications
Feb 08, 2022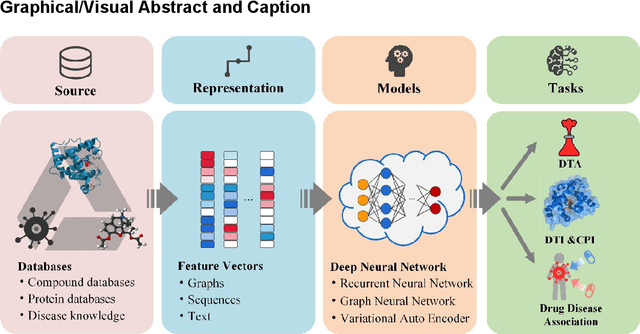
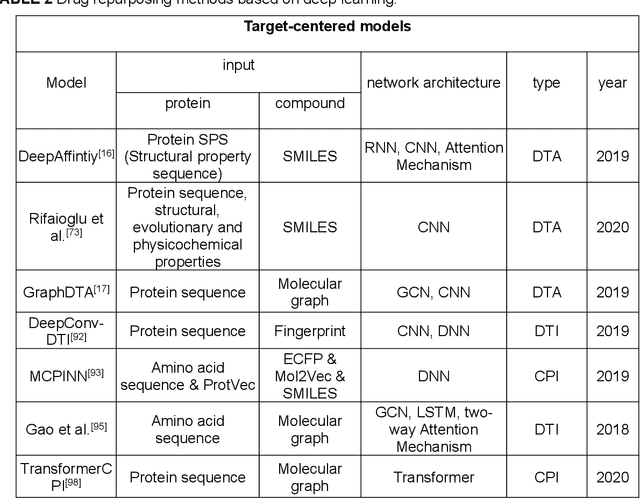
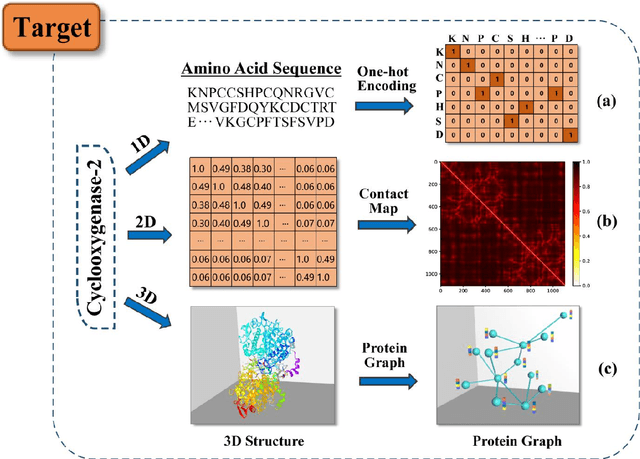
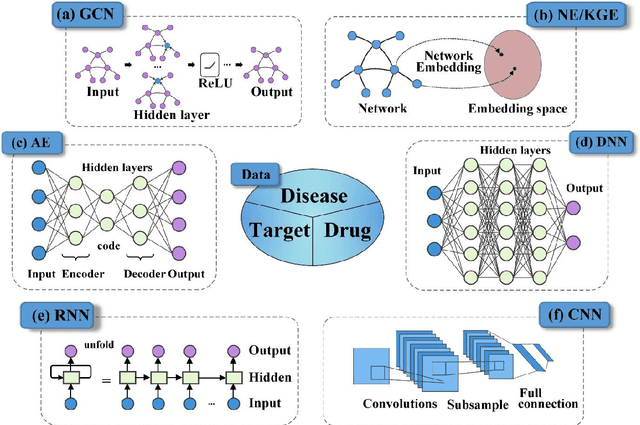
Abstract:Drug development is time-consuming and expensive. Repurposing existing drugs for new therapies is an attractive solution that accelerates drug development at reduced experimental costs, specifically for Coronavirus Disease 2019 (COVID-19), an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, comprehensively obtaining and productively integrating available knowledge and big biomedical data to effectively advance deep learning models is still challenging for drug repurposing in other complex diseases. In this review, we introduce guidelines on how to utilize deep learning methodologies and tools for drug repurposing. We first summarized the commonly used bioinformatics and pharmacogenomics databases for drug repurposing. Next, we discuss recently developed sequence-based and graph-based representation approaches as well as state-of-the-art deep learning-based methods. Finally, we present applications of drug repurposing to fight the COVID-19 pandemic, and outline its future challenges.
Repurpose Open Data to Discover Therapeutics for COVID-19 using Deep Learning
May 21, 2020Abstract:There have been more than 850,000 confirmed cases and over 48,000 deaths from the human coronavirus disease 2019 (COVID-19) pandemic, caused by novel severe acute respiratory syndrome coronavirus (SARS-CoV-2), in the United States alone. However, there are currently no proven effective medications against COVID-19. Drug repurposing offers a promising way for the development of prevention and treatment strategies for COVID-19. This study reports an integrative, network-based deep learning methodology to identify repurposable drugs for COVID-19 (termed CoV-KGE). Specifically, we built a comprehensive knowledge graph that includes 15 million edges across 39 types of relationships connecting drugs, diseases, genes, pathways, and expressions, from a large scientific corpus of 24 million PubMed publications. Using Amazon AWS computing resources, we identified 41 repurposable drugs (including indomethacin, toremifene and niclosamide) whose therapeutic association with COVID-19 were validated by transcriptomic and proteomic data in SARS-CoV-2 infected human cells and data from ongoing clinical trials. While this study, by no means recommends specific drugs, it demonstrates a powerful deep learning methodology to prioritize existing drugs for further investigation, which holds the potential of accelerating therapeutic development for COVID-19.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge