Jiajing Hu
DGL-LifeSci: An Open-Source Toolkit for Deep Learning on Graphs in Life Science
Jun 27, 2021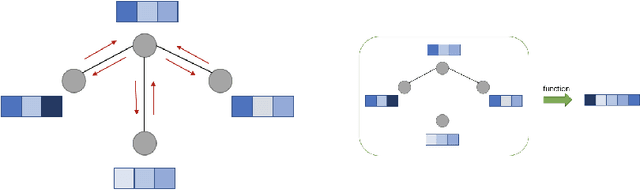
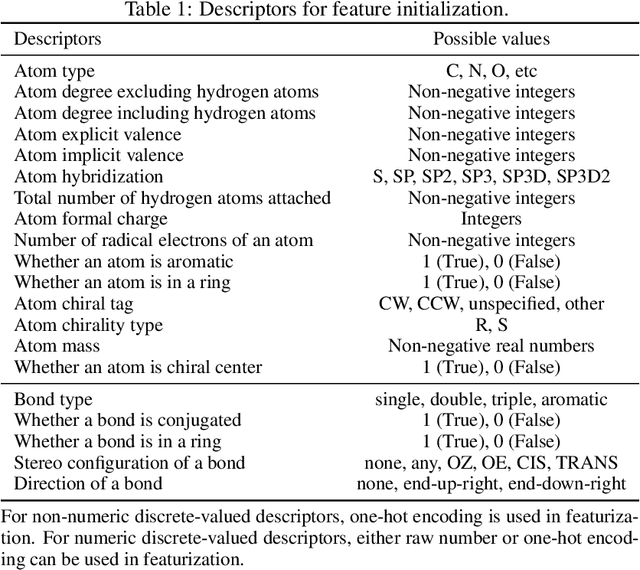

Abstract:Graph neural networks (GNNs) constitute a class of deep learning methods for graph data. They have wide applications in chemistry and biology, such as molecular property prediction, reaction prediction and drug-target interaction prediction. Despite the interest, GNN-based modeling is challenging as it requires graph data pre-processing and modeling in addition to programming and deep learning. Here we present DGL-LifeSci, an open-source package for deep learning on graphs in life science. DGL-LifeSci is a python toolkit based on RDKit, PyTorch and Deep Graph Library (DGL). DGL-LifeSci allows GNN-based modeling on custom datasets for molecular property prediction, reaction prediction and molecule generation. With its command-line interfaces, users can perform modeling without any background in programming and deep learning. We test the command-line interfaces using standard benchmarks MoleculeNet, USPTO, and ZINC. Compared with previous implementations, DGL-LifeSci achieves a speed up by up to 6x. For modeling flexibility, DGL-LifeSci provides well-optimized modules for various stages of the modeling pipeline. In addition, DGL-LifeSci provides pre-trained models for reproducing the test experiment results and applying models without training. The code is distributed under an Apache-2.0 License and is freely accessible at https://github.com/awslabs/dgl-lifesci.
Order Matters: Probabilistic Modeling of Node Sequence for Graph Generation
Jun 14, 2021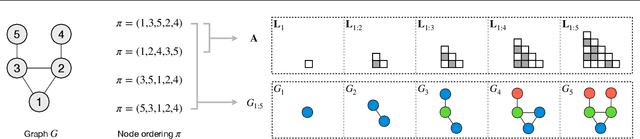
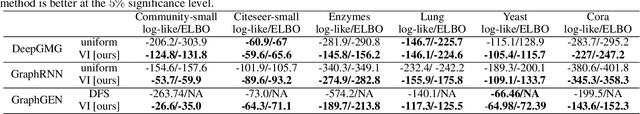
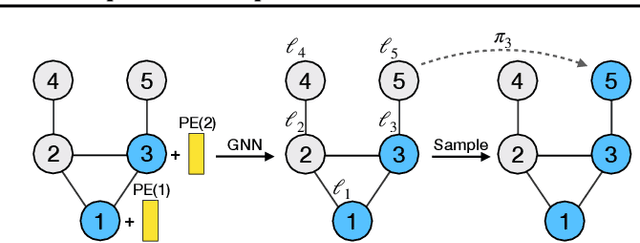
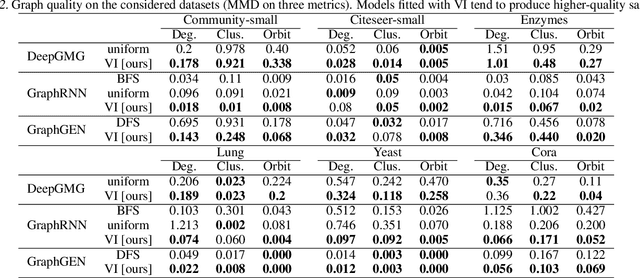
Abstract:A graph generative model defines a distribution over graphs. One type of generative model is constructed by autoregressive neural networks, which sequentially add nodes and edges to generate a graph. However, the likelihood of a graph under the autoregressive model is intractable, as there are numerous sequences leading to the given graph; this makes maximum likelihood estimation challenging. Instead, in this work we derive the exact joint probability over the graph and the node ordering of the sequential process. From the joint, we approximately marginalize out the node orderings and compute a lower bound on the log-likelihood using variational inference. We train graph generative models by maximizing this bound, without using the ad-hoc node orderings of previous methods. Our experiments show that the log-likelihood bound is significantly tighter than the bound of previous schemes. Moreover, the models fitted with the proposed algorithm can generate high-quality graphs that match the structures of target graphs not seen during training. We have made our code publicly available at \hyperref[https://github.com/tufts-ml/graph-generation-vi]{https://github.com/tufts-ml/graph-generation-vi}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge