Xiaohui Chen
Hierarchy of discriminative power and complexity in learning quantum ensembles
Jan 29, 2026Abstract:Distance metrics are central to machine learning, yet distances between ensembles of quantum states remain poorly understood due to fundamental quantum measurement constraints. We introduce a hierarchy of integral probability metrics, termed MMD-$k$, which generalizes the maximum mean discrepancy to quantum ensembles and exhibit a strict trade-off between discriminative power and statistical efficiency as the moment order $k$ increases. For pure-state ensembles of size $N$, estimating MMD-$k$ using experimentally feasible SWAP-test-based estimators requires $Θ(N^{2-2/k})$ samples for constant $k$, and $Θ(N^3)$ samples to achieve full discriminative power at $k = N$. In contrast, the quantum Wasserstein distance attains full discriminative power with $Θ(N^2 \log N)$ samples. These results provide principled guidance for the design of loss functions in quantum machine learning, which we illustrate in the training quantum denoising diffusion probabilistic models.
SpecBridge: Bridging Mass Spectrometry and Molecular Representations via Cross-Modal Alignment
Jan 27, 2026Abstract:Small-molecule identification from tandem mass spectrometry (MS/MS) remains a bottleneck in untargeted settings where spectral libraries are incomplete. While deep learning offers a solution, current approaches typically fall into two extremes: explicit generative models that construct molecular graphs atom-by-atom, or joint contrastive models that learn cross-modal subspaces from scratch. We introduce SpecBridge, a novel implicit alignment framework that treats structure identification as a geometric alignment problem. SpecBridge fine-tunes a self-supervised spectral encoder (DreaMS) to project directly into the latent space of a frozen molecular foundation model (ChemBERTa), and then performs retrieval by cosine similarity to a fixed bank of precomputed molecular embeddings. Across MassSpecGym, Spectraverse, and MSnLib benchmarks, SpecBridge improves top-1 retrieval accuracy by roughly 20-25% relative to strong neural baselines, while keeping the number of trainable parameters small. These results suggest that aligning to frozen foundation models is a practical, stable alternative to designing new architectures from scratch. The code for SpecBridge is released at https://github.com/HassounLab/SpecBridge.
OpenPathNet: An Open-Source RF Multipath Data Generator for AI-Driven Wireless Systems
Dec 19, 2025Abstract:The convergence of artificial intelligence (AI) and sixth-generation (6G) wireless technologies is driving an urgent need for large-scale, high-fidelity, and reproducible radio frequency (RF) datasets. Existing resources, such as CKMImageNet, primarily provide preprocessed and image-based channel representations, which conceal the fine-grained physical characteristics of signal propagation that are essential for effective AI modeling. To bridge this gap, we present OpenPathNet, an open-source RF multipath data generator accompanied by a publicly released dataset for AI-driven wireless research. Distinct from prior datasets, OpenPathNet offers disaggregated and physically consistent multipath parameters, including per-path gain, time of arrival (ToA), and spatial angles, derived from high-precision ray tracing simulations constructed on real-world environment maps. By adopting a modular, parameterized pipeline, OpenPathNet enables reproducible generation of multipath data and can be readily extended to new environments and configurations, improving scalability and transparency. The released generator and accompanying dataset provide an extensible testbed that holds promise for advancing studies on channel modeling, beam prediction, environment-aware communication, and integrated sensing in AI-enabled 6G systems. The source code and dataset are publicly available at https://github.com/liu-lz/OpenPathNet.
Neural Local Wasserstein Regression
Nov 13, 2025Abstract:We study the estimation problem of distribution-on-distribution regression, where both predictors and responses are probability measures. Existing approaches typically rely on a global optimal transport map or tangent-space linearization, which can be restrictive in approximation capacity and distort geometry in multivariate underlying domains. In this paper, we propose the \emph{Neural Local Wasserstein Regression}, a flexible nonparametric framework that models regression through locally defined transport maps in Wasserstein space. Our method builds on the analogy with classical kernel regression: kernel weights based on the 2-Wasserstein distance localize estimators around reference measures, while neural networks parameterize transport operators that adapt flexibly to complex data geometries. This localized perspective broadens the class of admissible transformations and avoids the limitations of global map assumptions and linearization structures. We develop a practical training procedure using DeepSets-style architectures and Sinkhorn-approximated losses, combined with a greedy reference selection strategy for scalability. Through synthetic experiments on Gaussian and mixture models, as well as distributional prediction tasks on MNIST, we demonstrate that our approach effectively captures nonlinear and high-dimensional distributional relationships that elude existing methods.
PINN and GNN-based RF Map Construction for Wireless Communication Systems
Jul 30, 2025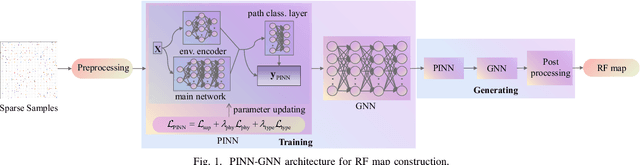
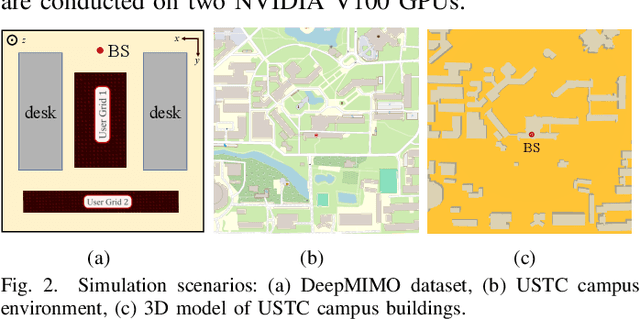
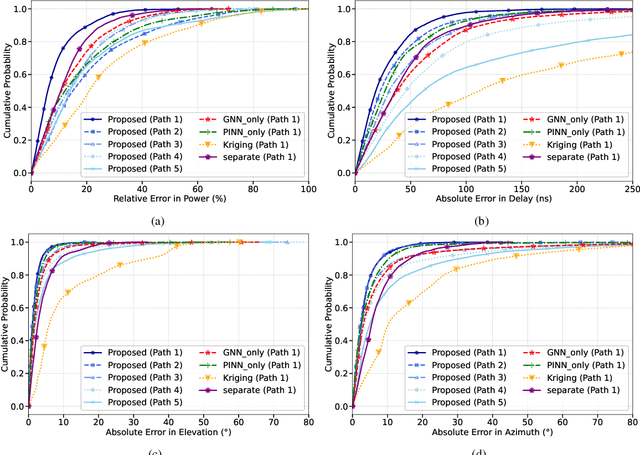
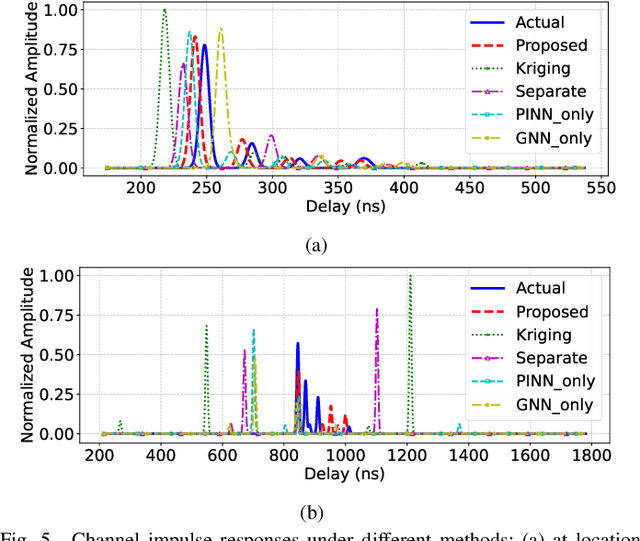
Abstract:Radio frequency (RF) map is a promising technique for capturing the characteristics of multipath signal propagation, offering critical support for channel modeling, coverage analysis, and beamforming in wireless communication networks. This paper proposes a novel RF map construction method based on a combination of physics-informed neural network (PINN) and graph neural network (GNN). The PINN incorporates physical constraints derived from electromagnetic propagation laws to guide the learning process, while the GNN models spatial correlations among receiver locations. By parameterizing multipath signals into received power, delay, and angle of arrival (AoA), and integrating both physical priors and spatial dependencies, the proposed method achieves accurate prediction of multipath parameters. Experimental results demonstrate that the method enables high-precision RF map construction under sparse sampling conditions and delivers robust performance in both indoor and complex outdoor environments, outperforming baseline methods in terms of generalization and accuracy.
A Generalized Label Shift Perspective for Cross-Domain Gaze Estimation
May 19, 2025Abstract:Aiming to generalize the well-trained gaze estimation model to new target domains, Cross-domain Gaze Estimation (CDGE) is developed for real-world application scenarios. Existing CDGE methods typically extract the domain-invariant features to mitigate domain shift in feature space, which is proved insufficient by Generalized Label Shift (GLS) theory. In this paper, we introduce a novel GLS perspective to CDGE and modelize the cross-domain problem by label and conditional shift problem. A GLS correction framework is presented and a feasible realization is proposed, in which a importance reweighting strategy based on truncated Gaussian distribution is introduced to overcome the continuity challenges in label shift correction. To embed the reweighted source distribution to conditional invariant learning, we further derive a probability-aware estimation of conditional operator discrepancy. Extensive experiments on standard CDGE tasks with different backbone models validate the superior generalization capability across domain and applicability on various models of proposed method.
Sobolev Gradient Ascent for Optimal Transport: Barycenter Optimization and Convergence Analysis
May 19, 2025Abstract:This paper introduces a new constraint-free concave dual formulation for the Wasserstein barycenter. Tailoring the vanilla dual gradient ascent algorithm to the Sobolev geometry, we derive a scalable Sobolev gradient ascent (SGA) algorithm to compute the barycenter for input distributions supported on a regular grid. Despite the algorithmic simplicity, we provide a global convergence analysis that achieves the same rate as the classical subgradient descent methods for minimizing nonsmooth convex functions in the Euclidean space. A central feature of our SGA algorithm is that the computationally expensive $c$-concavity projection operator enforced on the Kantorovich dual potentials is unnecessary to guarantee convergence, leading to significant algorithmic and theoretical simplifications over all existing primal and dual methods for computing the exact barycenter. Our numerical experiments demonstrate the superior empirical performance of SGA over the existing optimal transport barycenter solvers.
Embedding Empirical Distributions for Computing Optimal Transport Maps
Apr 24, 2025Abstract:Distributional data have become increasingly prominent in modern signal processing, highlighting the necessity of computing optimal transport (OT) maps across multiple probability distributions. Nevertheless, recent studies on neural OT methods predominantly focused on the efficient computation of a single map between two distributions. To address this challenge, we introduce a novel approach to learning transport maps for new empirical distributions. Specifically, we employ the transformer architecture to produce embeddings from distributional data of varying length; these embeddings are then fed into a hypernetwork to generate neural OT maps. Various numerical experiments were conducted to validate the embeddings and the generated OT maps. The model implementation and the code are provided on https://github.com/jiangmingchen/HOTET.
Multi-Modal Foundation Models for Computational Pathology: A Survey
Mar 12, 2025



Abstract:Foundation models have emerged as a powerful paradigm in computational pathology (CPath), enabling scalable and generalizable analysis of histopathological images. While early developments centered on uni-modal models trained solely on visual data, recent advances have highlighted the promise of multi-modal foundation models that integrate heterogeneous data sources such as textual reports, structured domain knowledge, and molecular profiles. In this survey, we provide a comprehensive and up-to-date review of multi-modal foundation models in CPath, with a particular focus on models built upon hematoxylin and eosin (H&E) stained whole slide images (WSIs) and tile-level representations. We categorize 32 state-of-the-art multi-modal foundation models into three major paradigms: vision-language, vision-knowledge graph, and vision-gene expression. We further divide vision-language models into non-LLM-based and LLM-based approaches. Additionally, we analyze 28 available multi-modal datasets tailored for pathology, grouped into image-text pairs, instruction datasets, and image-other modality pairs. Our survey also presents a taxonomy of downstream tasks, highlights training and evaluation strategies, and identifies key challenges and future directions. We aim for this survey to serve as a valuable resource for researchers and practitioners working at the intersection of pathology and AI.
High-fidelity Multiphysics Modelling for Rapid Predictions Using Physics-informed Parallel Neural Operator
Feb 26, 2025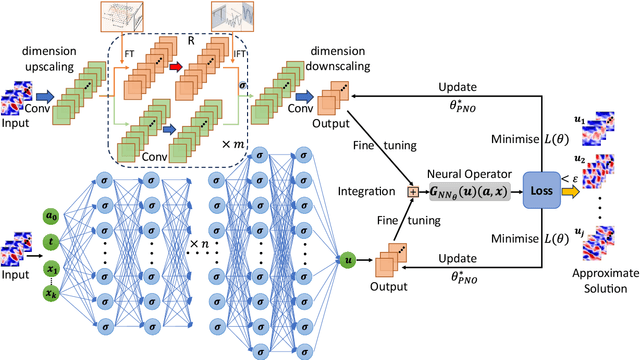
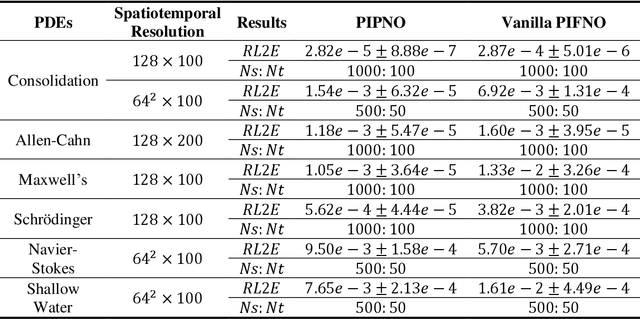
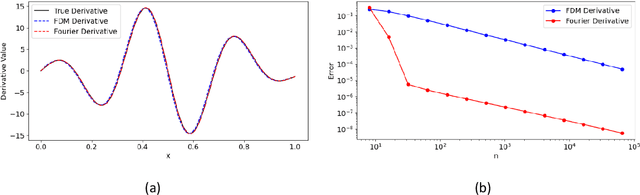
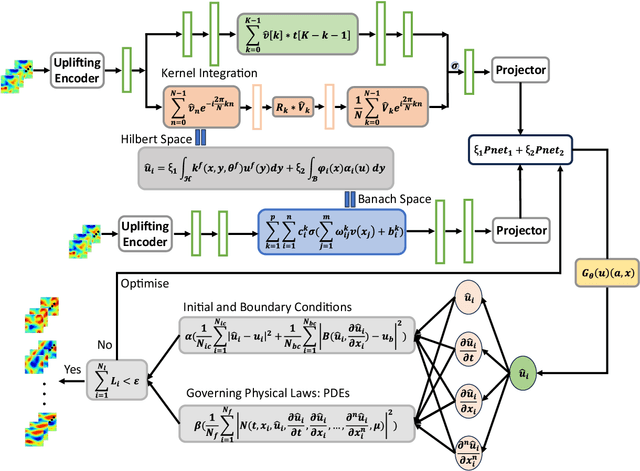
Abstract:Modelling complex multiphysics systems governed by nonlinear and strongly coupled partial differential equations (PDEs) is a cornerstone in computational science and engineering. However, it remains a formidable challenge for traditional numerical solvers due to high computational cost, making them impractical for large-scale applications. Neural operators' reliance on data-driven training limits their applicability in real-world scenarios, as data is often scarce or expensive to obtain. Here, we propose a novel paradigm, physics-informed parallel neural operator (PIPNO), a scalable and unsupervised learning framework that enables data-free PDE modelling by leveraging only governing physical laws. The parallel kernel integration design, incorporating ensemble learning, significantly enhances both compatibility and computational efficiency, enabling scalable operator learning for nonlinear and strongly coupled PDEs. PIPNO efficiently captures nonlinear operator mappings across diverse physics, including geotechnical engineering, material science, electromagnetism, quantum mechanics, and fluid dynamics. The proposed method achieves high-fidelity and rapid predictions, outperforming existing operator learning approaches in modelling nonlinear and strongly coupled multiphysics systems. Therefore, PIPNO offers a powerful alternative to conventional solvers, broadening the applicability of neural operators for multiphysics modelling while ensuring efficiency, robustness, and scalability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge