Jianan Canal Li
A Phylogenetic Approach to Genomic Language Modeling
Mar 04, 2025



Abstract:Genomic language models (gLMs) have shown mostly modest success in identifying evolutionarily constrained elements in mammalian genomes. To address this issue, we introduce a novel framework for training gLMs that explicitly models nucleotide evolution on phylogenetic trees using multispecies whole-genome alignments. Our approach integrates an alignment into the loss function during training but does not require it for making predictions, thereby enhancing the model's applicability. We applied this framework to train PhyloGPN, a model that excels at predicting functionally disruptive variants from a single sequence alone and demonstrates strong transfer learning capabilities.
Large Language Model is Secretly a Protein Sequence Optimizer
Jan 16, 2025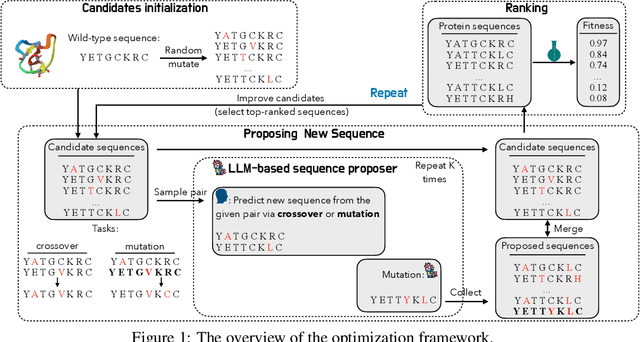
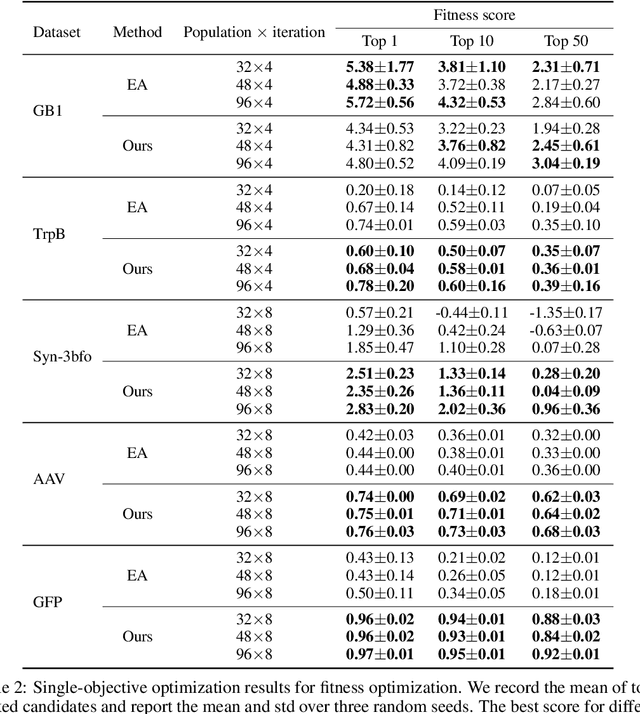
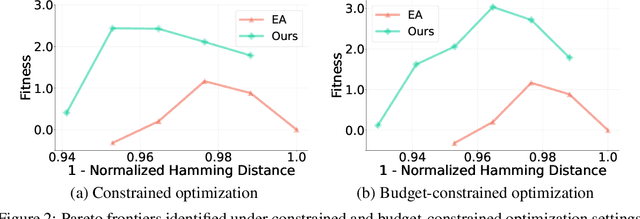
Abstract:We consider the protein sequence engineering problem, which aims to find protein sequences with high fitness levels, starting from a given wild-type sequence. Directed evolution has been a dominating paradigm in this field which has an iterative process to generate variants and select via experimental feedback. We demonstrate large language models (LLMs), despite being trained on massive texts, are secretly protein sequence optimizers. With a directed evolutionary method, LLM can perform protein engineering through Pareto and experiment-budget constrained optimization, demonstrating success on both synthetic and experimental fitness landscapes.
Genomic Language Models: Opportunities and Challenges
Jul 16, 2024



Abstract:Large language models (LLMs) are having transformative impacts across a wide range of scientific fields, particularly in the biomedical sciences. Just as the goal of Natural Language Processing is to understand sequences of words, a major objective in biology is to understand biological sequences. Genomic Language Models (gLMs), which are LLMs trained on DNA sequences, have the potential to significantly advance our understanding of genomes and how DNA elements at various scales interact to give rise to complex functions. In this review, we showcase this potential by highlighting key applications of gLMs, including fitness prediction, sequence design, and transfer learning. Despite notable recent progress, however, developing effective and efficient gLMs presents numerous challenges, especially for species with large, complex genomes. We discuss major considerations for developing and evaluating gLMs.
MCTensor: A High-Precision Deep Learning Library with Multi-Component Floating-Point
Jul 18, 2022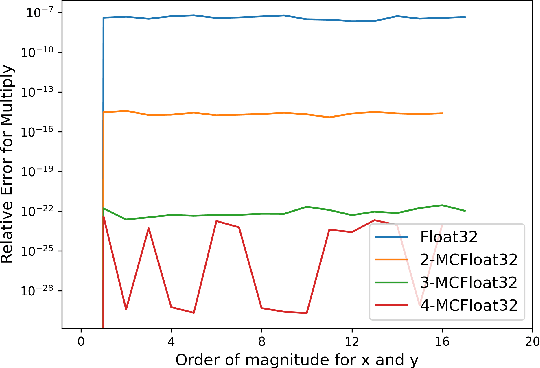

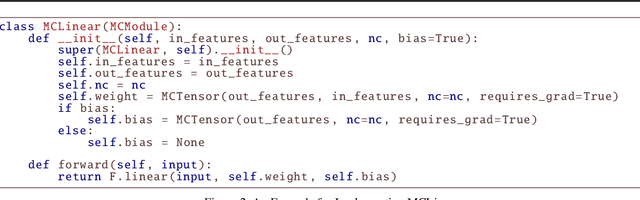
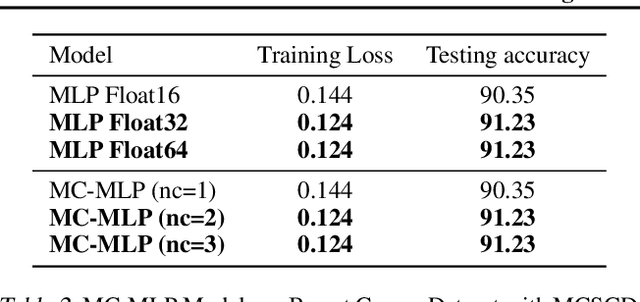
Abstract:In this paper, we introduce MCTensor, a library based on PyTorch for providing general-purpose and high-precision arithmetic for DL training. MCTensor is used in the same way as PyTorch Tensor: we implement multiple basic, matrix-level computation operators and NN modules for MCTensor with identical PyTorch interface. Our algorithms achieve high precision computation and also benefits from heavily-optimized PyTorch floating-point arithmetic. We evaluate MCTensor arithmetic against PyTorch native arithmetic for a series of tasks, where models using MCTensor in float16 would match or outperform the PyTorch model with float32 or float64 precision.
Cyclical Kernel Adaptive Metropolis
Jun 30, 2022


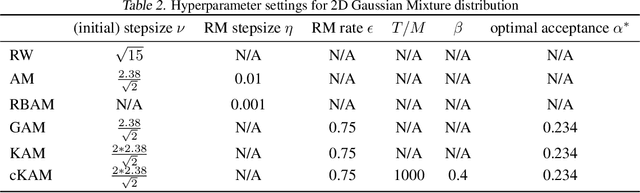
Abstract:We propose cKAM, cyclical Kernel Adaptive Metropolis, which incorporates a cyclical stepsize scheme to allow control for exploration and sampling. We show that on a crafted bimodal distribution, existing Adaptive Metropolis type algorithms would fail to converge to the true posterior distribution. We point out that this is because adaptive samplers estimates the local/global covariance structure using past history of the chain, which will lead to adaptive algorithms be trapped in a local mode. We demonstrate that cKAM encourages exploration of the posterior distribution and allows the sampler to escape from a local mode, while maintaining the high performance of adaptive methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge