Liping Liu
Dept. of Computer Science, Tufts University
Instruct-of-Reflection: Enhancing Large Language Models Iterative Reflection Capabilities via Dynamic-Meta Instruction
Mar 02, 2025



Abstract:Self-reflection for Large Language Models (LLMs) has gained significant attention. Existing approaches involve models iterating and improving their previous responses based on LLMs' internal reflection ability or external feedback. However, recent research has raised doubts about whether intrinsic self-correction without external feedback may even degrade performance. Based on our empirical evidence, we find that current static reflection methods may lead to redundant, drift, and stubborn issues. To mitigate this, we introduce Instruct-of-Reflection (IoRT), a novel and general reflection framework that leverages dynamic-meta instruction to enhance the iterative reflection capability of LLMs. Specifically, we propose the instructor driven by the meta-thoughts and self-consistency classifier, generates various instructions, including refresh, stop, and select, to guide the next reflection iteration. Our experiments demonstrate that IoRT achieves an average improvement of 10.1% over established baselines in mathematical and commonsense reasoning tasks, highlighting its efficacy and applicability.
MADGEN -- Mass-Spec attends to De Novo Molecular generation
Jan 03, 2025Abstract:The annotation (assigning structural chemical identities) of MS/MS spectra remains a significant challenge due to the enormous molecular diversity in biological samples and the limited scope of reference databases. Currently, the vast majority of spectral measurements remain in the "dark chemical space" without structural annotations. To improve annotation, we propose MADGEN (Mass-spec Attends to De Novo Molecular GENeration), a scaffold-based method for de novo molecular structure generation guided by mass spectrometry data. MADGEN operates in two stages: scaffold retrieval and spectra-conditioned molecular generation starting with the scaffold. In the first stage, given an MS/MS spectrum, we formulate scaffold retrieval as a ranking problem and employ contrastive learning to align mass spectra with candidate molecular scaffolds. In the second stage, starting from the retrieved scaffold, we employ the MS/MS spectrum to guide an attention-based generative model to generate the final molecule. Our approach constrains the molecular generation search space, reducing its complexity and improving generation accuracy. We evaluate MADGEN on three datasets (NIST23, CANOPUS, and MassSpecGym) and evaluate MADGEN's performance with a predictive scaffold retriever and with an oracle retriever. We demonstrate the effectiveness of using attention to integrate spectral information throughout the generation process to achieve strong results with the oracle retriever.
Enhancing Diffusion-based Point Cloud Generation with Smoothness Constraint
Apr 03, 2024Abstract:Diffusion models have been popular for point cloud generation tasks. Existing works utilize the forward diffusion process to convert the original point distribution into a noise distribution and then learn the reverse diffusion process to recover the point distribution from the noise distribution. However, the reverse diffusion process can produce samples with non-smooth points on the surface because of the ignorance of the point cloud geometric properties. We propose alleviating the problem by incorporating the local smoothness constraint into the diffusion framework for point cloud generation. Experiments demonstrate the proposed model can generate realistic shapes and smoother point clouds, outperforming multiple state-of-the-art methods.
Graph Pruning for Enumeration of Minimal Unsatisfiable Subsets
Feb 19, 2024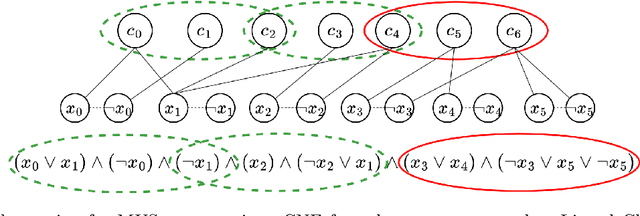
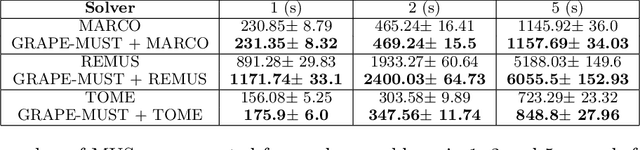
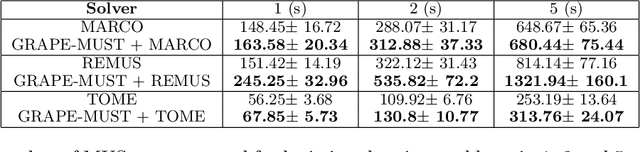

Abstract:Finding Minimal Unsatisfiable Subsets (MUSes) of binary constraints is a common problem in infeasibility analysis of over-constrained systems. However, because of the exponential search space of the problem, enumerating MUSes is extremely time-consuming in real applications. In this work, we propose to prune formulas using a learned model to speed up MUS enumeration. We represent formulas as graphs and then develop a graph-based learning model to predict which part of the formula should be pruned. Importantly, our algorithm does not require data labeling by only checking the satisfiability of pruned formulas. It does not even require training data from the target application because it extrapolates to data with different distributions. In our experiments we combine our algorithm with existing MUS enumerators and validate its effectiveness in multiple benchmarks including a set of real-world problems outside our training distribution. The experiment results show that our method significantly accelerates MUS enumeration on average on these benchmark problems.
Development of a Deep Learning System for Intra-Operative Identification of Cancer Metastases
Jun 17, 2023Abstract:For several cancer patients, operative resection with curative intent can end up in early recurrence of the cancer. Current limitations in peri-operative cancer staging and especially intra-operative misidentification of visible metastases is likely the main reason leading to unnecessary operative interventions in the affected individuals. Here, we evaluate whether an artificial intelligence (AI) system can improve recognition of peritoneal surface metastases on routine staging laparoscopy images from patients with gastrointestinal malignancies. In a simulated setting evaluating biopsied peritoneal lesions, a prototype deep learning surgical guidance system outperformed oncologic surgeons in identifying peritoneal surface metastases. In this environment the developed AI model would have improved the identification of metastases by 5% while reducing the number of unnecessary biopsies by 28% compared to current standard practice. Evaluating non-biopsied peritoneal lesions, the findings support the possibility that the AI system could identify peritoneal surface metastases that were falsely deemed benign in clinical practice. Our findings demonstrate the technical feasibility of an AI system for intra-operative identification of peritoneal surface metastases, but require future assessment in a multi-institutional clinical setting.
Interpretable Node Representation with Attribute Decoding
Dec 03, 2022Abstract:Variational Graph Autoencoders (VGAEs) are powerful models for unsupervised learning of node representations from graph data. In this work, we systematically analyze modeling node attributes in VGAEs and show that attribute decoding is important for node representation learning. We further propose a new learning model, interpretable NOde Representation with Attribute Decoding (NORAD). The model encodes node representations in an interpretable approach: node representations capture community structures in the graph and the relationship between communities and node attributes. We further propose a rectifying procedure to refine node representations of isolated notes, improving the quality of these nodes' representations. Our empirical results demonstrate the advantage of the proposed model when learning graph data in an interpretable approach.
NovelCraft: A Dataset for Novelty Detection and Discovery in Open Worlds
Jun 23, 2022
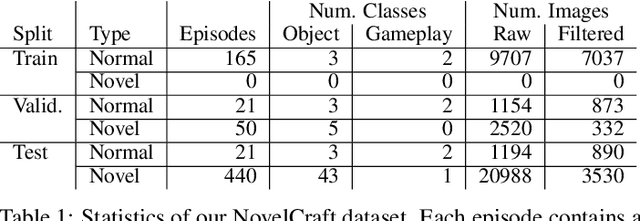
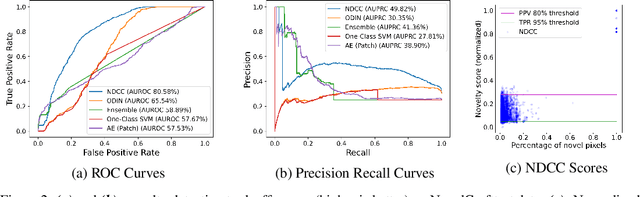
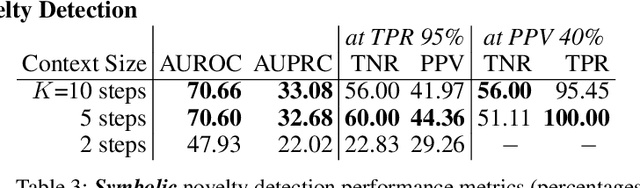
Abstract:In order for artificial agents to perform useful tasks in changing environments, they must be able to both detect and adapt to novelty. However, visual novelty detection research often only evaluates on repurposed datasets such as CIFAR-10 originally intended for object classification. This practice restricts novelties to well-framed images of distinct object types. We suggest that new benchmarks are needed to represent the challenges of navigating an open world. Our new NovelCraft dataset contains multi-modal episodic data of the images and symbolic world-states seen by an agent completing a pogo-stick assembly task within a video game world. In some episodes, we insert novel objects that can impact gameplay. Novelty can vary in size, position, and occlusion within complex scenes. We benchmark state-of-the-art novelty detection and generalized category discovery models with a focus on comprehensive evaluation. Results suggest an opportunity for future research: models aware of task-specific costs of different types of mistakes could more effectively detect and adapt to novelty in open worlds.
Order Matters: Probabilistic Modeling of Node Sequence for Graph Generation
Jun 14, 2021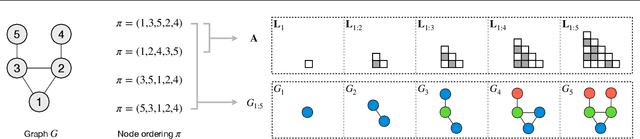
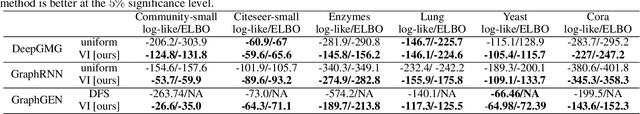
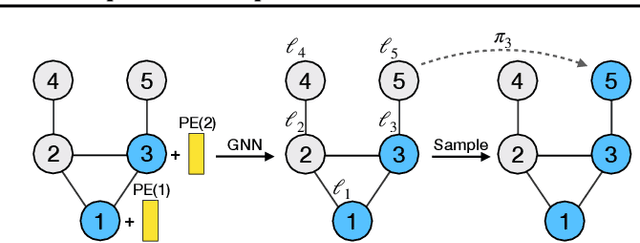
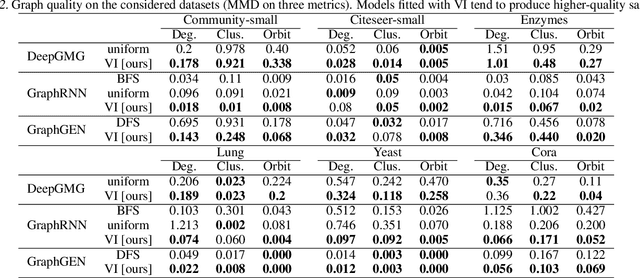
Abstract:A graph generative model defines a distribution over graphs. One type of generative model is constructed by autoregressive neural networks, which sequentially add nodes and edges to generate a graph. However, the likelihood of a graph under the autoregressive model is intractable, as there are numerous sequences leading to the given graph; this makes maximum likelihood estimation challenging. Instead, in this work we derive the exact joint probability over the graph and the node ordering of the sequential process. From the joint, we approximately marginalize out the node orderings and compute a lower bound on the log-likelihood using variational inference. We train graph generative models by maximizing this bound, without using the ad-hoc node orderings of previous methods. Our experiments show that the log-likelihood bound is significantly tighter than the bound of previous schemes. Moreover, the models fitted with the proposed algorithm can generate high-quality graphs that match the structures of target graphs not seen during training. We have made our code publicly available at \hyperref[https://github.com/tufts-ml/graph-generation-vi]{https://github.com/tufts-ml/graph-generation-vi}.
DiaKG: an Annotated Diabetes Dataset for Medical Knowledge Graph Construction
May 31, 2021
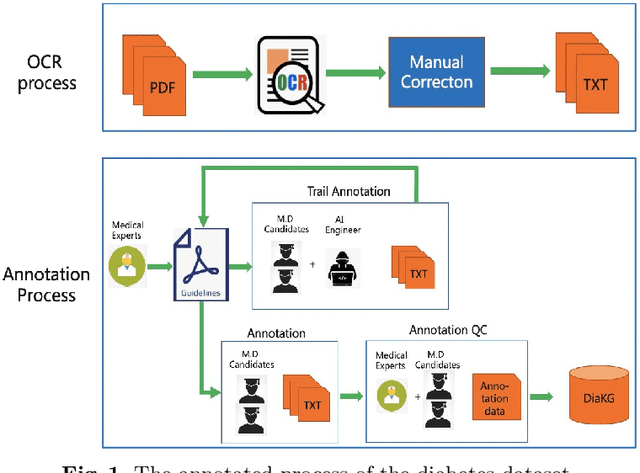

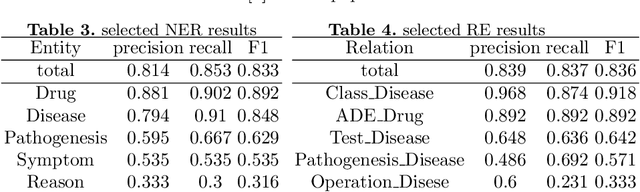
Abstract:Knowledge Graph has been proven effective in modeling structured information and conceptual knowledge, especially in the medical domain. However, the lack of high-quality annotated corpora remains a crucial problem for advancing the research and applications on this task. In order to accelerate the research for domain-specific knowledge graphs in the medical domain, we introduce DiaKG, a high-quality Chinese dataset for Diabetes knowledge graph, which contains 22,050 entities and 6,890 relations in total. We implement recent typical methods for Named Entity Recognition and Relation Extraction as a benchmark to evaluate the proposed dataset thoroughly. Empirical results show that the DiaKG is challenging for most existing methods and further analysis is conducted to discuss future research direction for improvements. We hope the release of this dataset can assist the construction of diabetes knowledge graphs and facilitate AI-based applications.
Using Graph Neural Networks for Mass Spectrometry Prediction
Oct 09, 2020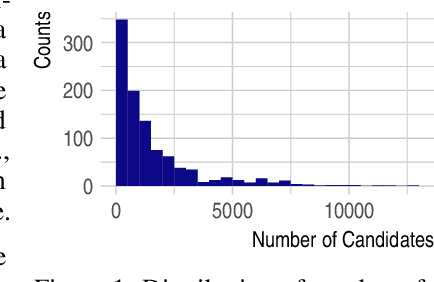
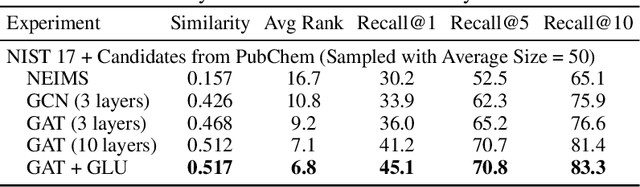
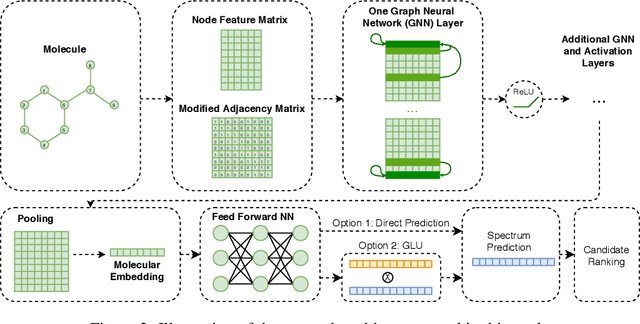

Abstract:Detecting and quantifying products of cellular metabolism using Mass Spectrometry (MS) has already shown great promise in many biological and biomedical applications. The biggest challenge in metabolomics is annotation, where measured spectra are assigned chemical identities. Despite advances, current methods provide limited annotation for measured spectra. Here, we explore using graph neural networks (GNNs) to predict the spectra. The input to our model is a molecular graph. The model is trained and tested on the NIST 17 LC-MS dataset. We compare our results to NEIMS, a neural network model that utilizes molecular fingerprints as inputs. Our results show that GNN-based models offer higher performance than NEIMS. Importantly, we show that ranking results heavily depend on the candidate set size and on the similarity of the candidates to the target molecule, thus highlighting the need for consistent, well-characterized evaluation protocols for this domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge