Peter K. Sorger
Laboratory of Systems Pharmacology, Harvard Medical School
Virtual Multiplex Staining for Histological Images using a Marker-wise Conditioned Diffusion Model
Aug 20, 2025Abstract:Multiplex imaging is revolutionizing pathology by enabling the simultaneous visualization of multiple biomarkers within tissue samples, providing molecular-level insights that traditional hematoxylin and eosin (H&E) staining cannot provide. However, the complexity and cost of multiplex data acquisition have hindered its widespread adoption. Additionally, most existing large repositories of H&E images lack corresponding multiplex images, limiting opportunities for multimodal analysis. To address these challenges, we leverage recent advances in latent diffusion models (LDMs), which excel at modeling complex data distributions utilizing their powerful priors for fine-tuning to a target domain. In this paper, we introduce a novel framework for virtual multiplex staining that utilizes pretrained LDM parameters to generate multiplex images from H&E images using a conditional diffusion model. Our approach enables marker-by-marker generation by conditioning the diffusion model on each marker, while sharing the same architecture across all markers. To tackle the challenge of varying pixel value distributions across different marker stains and to improve inference speed, we fine-tune the model for single-step sampling, enhancing both color contrast fidelity and inference efficiency through pixel-level loss functions. We validate our framework on two publicly available datasets, notably demonstrating its effectiveness in generating up to 18 different marker types with improved accuracy, a substantial increase over the 2-3 marker types achieved in previous approaches. This validation highlights the potential of our framework, pioneering virtual multiplex staining. Finally, this paper bridges the gap between H&E and multiplex imaging, potentially enabling retrospective studies and large-scale analyses of existing H&E image repositories.
Multi-Modal Foundation Models for Computational Pathology: A Survey
Mar 12, 2025



Abstract:Foundation models have emerged as a powerful paradigm in computational pathology (CPath), enabling scalable and generalizable analysis of histopathological images. While early developments centered on uni-modal models trained solely on visual data, recent advances have highlighted the promise of multi-modal foundation models that integrate heterogeneous data sources such as textual reports, structured domain knowledge, and molecular profiles. In this survey, we provide a comprehensive and up-to-date review of multi-modal foundation models in CPath, with a particular focus on models built upon hematoxylin and eosin (H&E) stained whole slide images (WSIs) and tile-level representations. We categorize 32 state-of-the-art multi-modal foundation models into three major paradigms: vision-language, vision-knowledge graph, and vision-gene expression. We further divide vision-language models into non-LLM-based and LLM-based approaches. Additionally, we analyze 28 available multi-modal datasets tailored for pathology, grouped into image-text pairs, instruction datasets, and image-other modality pairs. Our survey also presents a taxonomy of downstream tasks, highlights training and evaluation strategies, and identifies key challenges and future directions. We aim for this survey to serve as a valuable resource for researchers and practitioners working at the intersection of pathology and AI.
A Survey on Computational Pathology Foundation Models: Datasets, Adaptation Strategies, and Evaluation Tasks
Jan 27, 2025Abstract:Computational pathology foundation models (CPathFMs) have emerged as a powerful approach for analyzing histopathological data, leveraging self-supervised learning to extract robust feature representations from unlabeled whole-slide images. These models, categorized into uni-modal and multi-modal frameworks, have demonstrated promise in automating complex pathology tasks such as segmentation, classification, and biomarker discovery. However, the development of CPathFMs presents significant challenges, such as limited data accessibility, high variability across datasets, the necessity for domain-specific adaptation, and the lack of standardized evaluation benchmarks. This survey provides a comprehensive review of CPathFMs in computational pathology, focusing on datasets, adaptation strategies, and evaluation tasks. We analyze key techniques, such as contrastive learning and multi-modal integration, and highlight existing gaps in current research. Finally, we explore future directions from four perspectives for advancing CPathFMs. This survey serves as a valuable resource for researchers, clinicians, and AI practitioners, guiding the advancement of CPathFMs toward robust and clinically applicable AI-driven pathology solutions.
Scope2Screen: Focus+Context Techniques for Pathology Tumor Assessment in Multivariate Image Data
Oct 10, 2021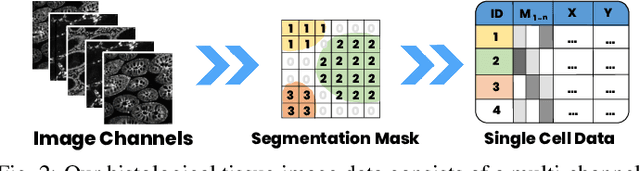

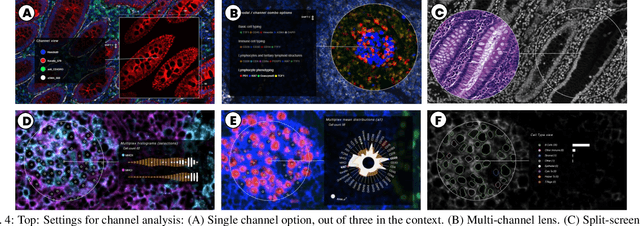
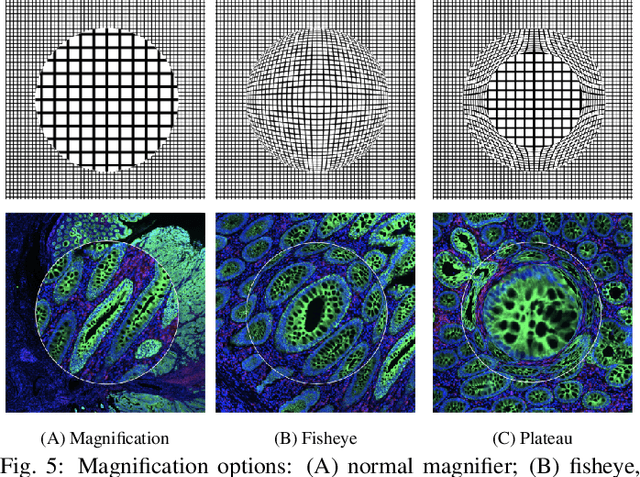
Abstract:Inspection of tissues using a light microscope is the primary method of diagnosing many diseases, notably cancer. Highly multiplexed tissue imaging builds on this foundation, enabling the collection of up to 60 channels of molecular information plus cell and tissue morphology using antibody staining. This provides unique insight into disease biology and promises to help with the design of patient-specific therapies. However, a substantial gap remains with respect to visualizing the resulting multivariate image data and effectively supporting pathology workflows in digital environments on screen. We, therefore, developed Scope2Screen, a scalable software system for focus+context exploration and annotation of whole-slide, high-plex, tissue images. Our approach scales to analyzing 100GB images of 10^9 or more pixels per channel, containing millions of cells. A multidisciplinary team of visualization experts, microscopists, and pathologists identified key image exploration and annotation tasks involving finding, magnifying, quantifying, and organizing ROIs in an intuitive and cohesive manner. Building on a scope2screen metaphor, we present interactive lensing techniques that operate at single-cell and tissue levels. Lenses are equipped with task-specific functionality and descriptive statistics, making it possible to analyze image features, cell types, and spatial arrangements (neighborhoods) across image channels and scales. A fast sliding-window search guides users to regions similar to those under the lens; these regions can be analyzed and considered either separately or as part of a larger image collection. A novel snapshot method enables linked lens configurations and image statistics to be saved, restored, and shared. We validate our designs with domain experts and apply Scope2Screen in two case studies involving lung and colorectal cancers to discover cancer-relevant image features.
MITI Minimum Information guidelines for highly multiplexed tissue images
Aug 21, 2021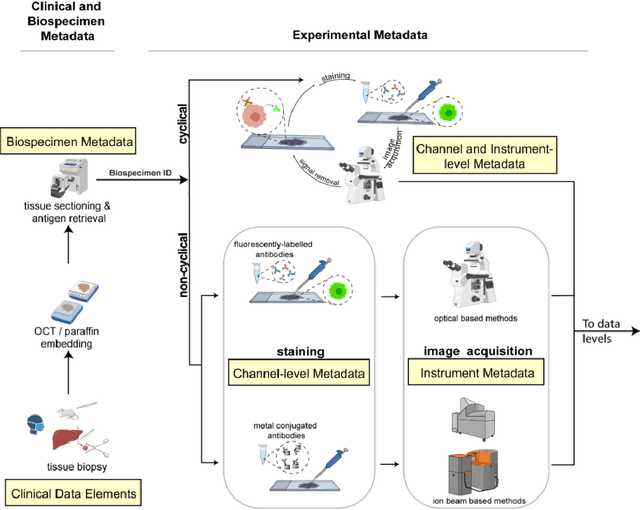
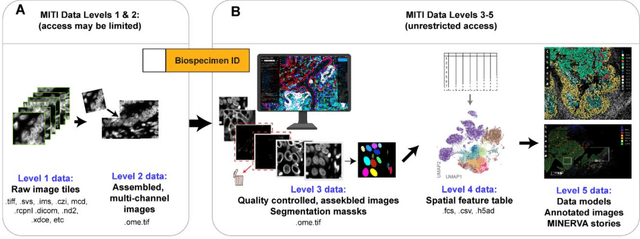
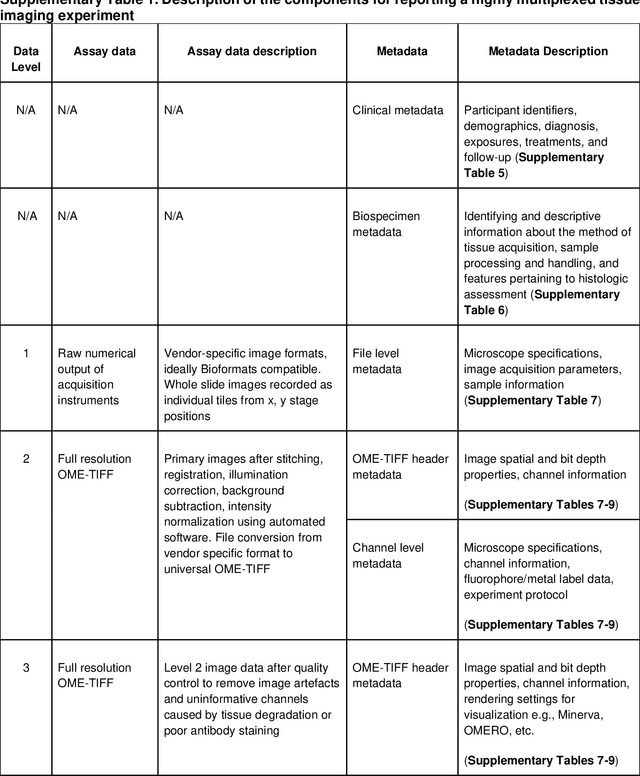
Abstract:The imminent release of atlases combining highly multiplexed tissue imaging with single cell sequencing and other omics data from human tissues and tumors creates an urgent need for data and metadata standards compliant with emerging and traditional approaches to histology. We describe the development of a Minimum Information about highly multiplexed Tissue Imaging (MITI) standard that draws on best practices from genomics and microscopy of cultured cells and model organisms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge