Zhiyi Shi
Adaptation of Agentic AI
Dec 22, 2025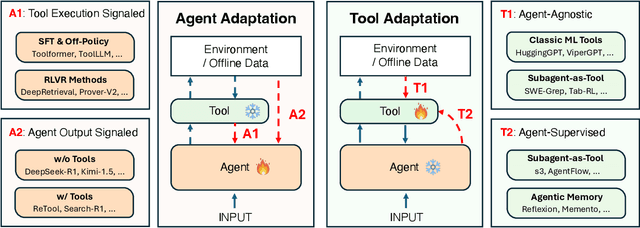
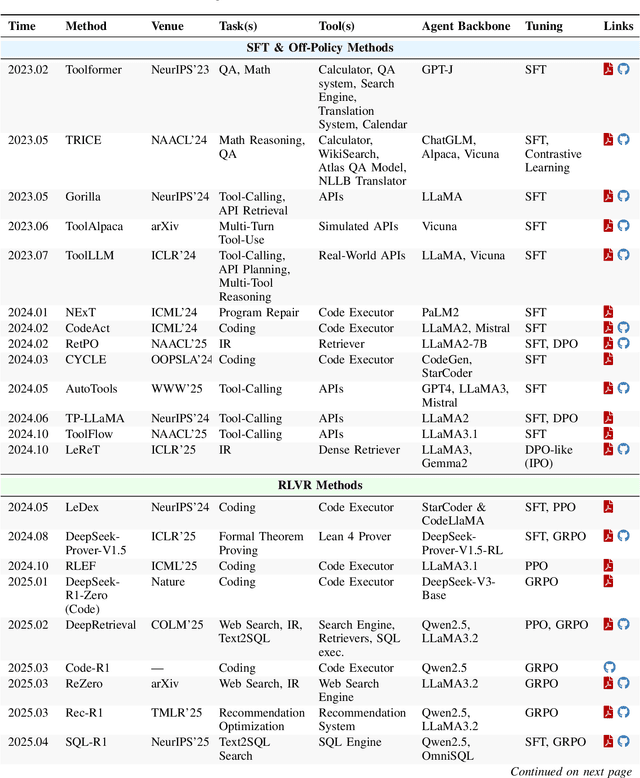
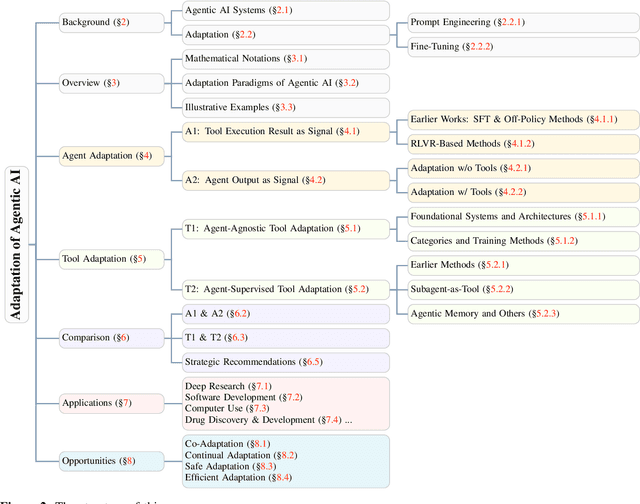
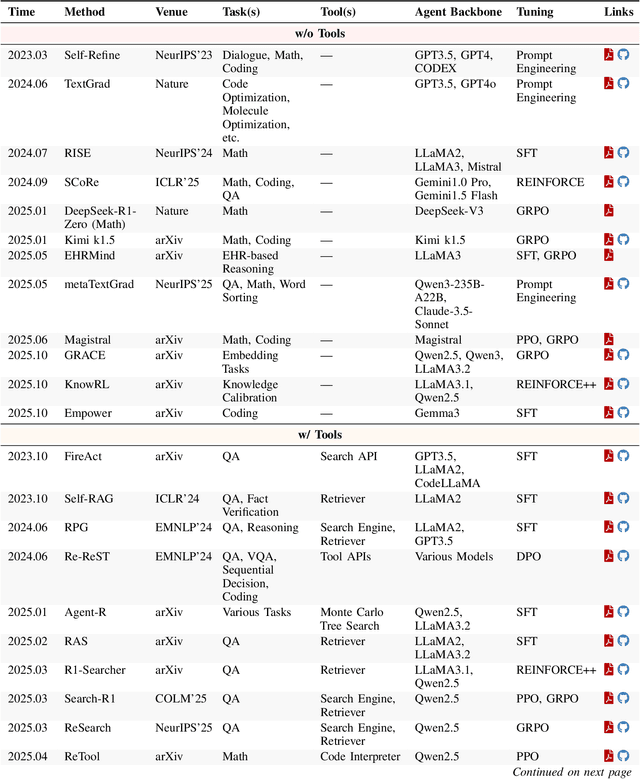
Abstract:Cutting-edge agentic AI systems are built on foundation models that can be adapted to plan, reason, and interact with external tools to perform increasingly complex and specialized tasks. As these systems grow in capability and scope, adaptation becomes a central mechanism for improving performance, reliability, and generalization. In this paper, we unify the rapidly expanding research landscape into a systematic framework that spans both agent adaptations and tool adaptations. We further decompose these into tool-execution-signaled and agent-output-signaled forms of agent adaptation, as well as agent-agnostic and agent-supervised forms of tool adaptation. We demonstrate that this framework helps clarify the design space of adaptation strategies in agentic AI, makes their trade-offs explicit, and provides practical guidance for selecting or switching among strategies during system design. We then review the representative approaches in each category, analyze their strengths and limitations, and highlight key open challenges and future opportunities. Overall, this paper aims to offer a conceptual foundation and practical roadmap for researchers and practitioners seeking to build more capable, efficient, and reliable agentic AI systems.
Virtual Multiplex Staining for Histological Images using a Marker-wise Conditioned Diffusion Model
Aug 20, 2025Abstract:Multiplex imaging is revolutionizing pathology by enabling the simultaneous visualization of multiple biomarkers within tissue samples, providing molecular-level insights that traditional hematoxylin and eosin (H&E) staining cannot provide. However, the complexity and cost of multiplex data acquisition have hindered its widespread adoption. Additionally, most existing large repositories of H&E images lack corresponding multiplex images, limiting opportunities for multimodal analysis. To address these challenges, we leverage recent advances in latent diffusion models (LDMs), which excel at modeling complex data distributions utilizing their powerful priors for fine-tuning to a target domain. In this paper, we introduce a novel framework for virtual multiplex staining that utilizes pretrained LDM parameters to generate multiplex images from H&E images using a conditional diffusion model. Our approach enables marker-by-marker generation by conditioning the diffusion model on each marker, while sharing the same architecture across all markers. To tackle the challenge of varying pixel value distributions across different marker stains and to improve inference speed, we fine-tune the model for single-step sampling, enhancing both color contrast fidelity and inference efficiency through pixel-level loss functions. We validate our framework on two publicly available datasets, notably demonstrating its effectiveness in generating up to 18 different marker types with improved accuracy, a substantial increase over the 2-3 marker types achieved in previous approaches. This validation highlights the potential of our framework, pioneering virtual multiplex staining. Finally, this paper bridges the gap between H&E and multiplex imaging, potentially enabling retrospective studies and large-scale analyses of existing H&E image repositories.
DualEdit: Dual Editing for Knowledge Updating in Vision-Language Models
Jun 16, 2025Abstract:Model editing aims to efficiently update a pre-trained model's knowledge without the need for time-consuming full retraining. While existing pioneering editing methods achieve promising results, they primarily focus on editing single-modal language models (LLMs). However, for vision-language models (VLMs), which involve multiple modalities, the role and impact of each modality on editing performance remain largely unexplored. To address this gap, we explore the impact of textual and visual modalities on model editing and find that: (1) textual and visual representations reach peak sensitivity at different layers, reflecting their varying importance; and (2) editing both modalities can efficiently update knowledge, but this comes at the cost of compromising the model's original capabilities. Based on our findings, we propose DualEdit, an editor that modifies both textual and visual modalities at their respective key layers. Additionally, we introduce a gating module within the more sensitive textual modality, allowing DualEdit to efficiently update new knowledge while preserving the model's original information. We evaluate DualEdit across multiple VLM backbones and benchmark datasets, demonstrating its superiority over state-of-the-art VLM editing baselines as well as adapted LLM editing methods on different evaluation metrics.
MAP: Revisiting Weight Decomposition for Low-Rank Adaptation
May 29, 2025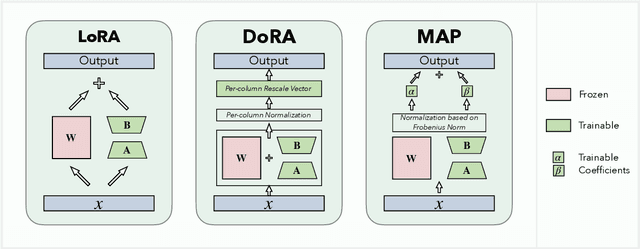
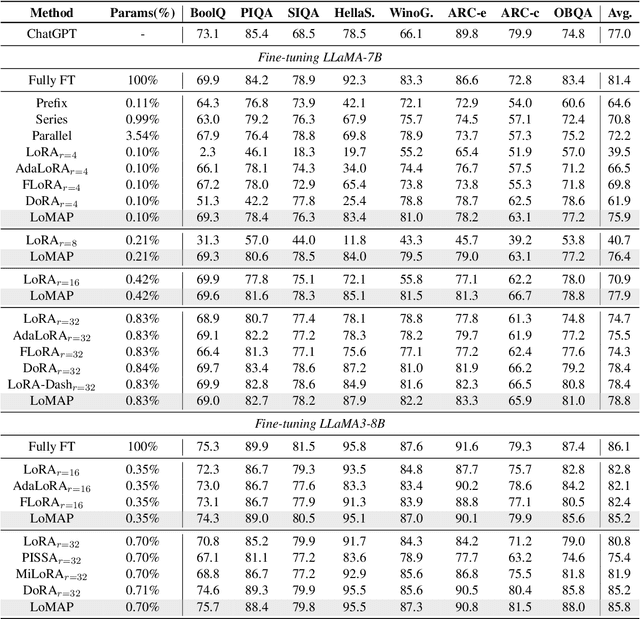
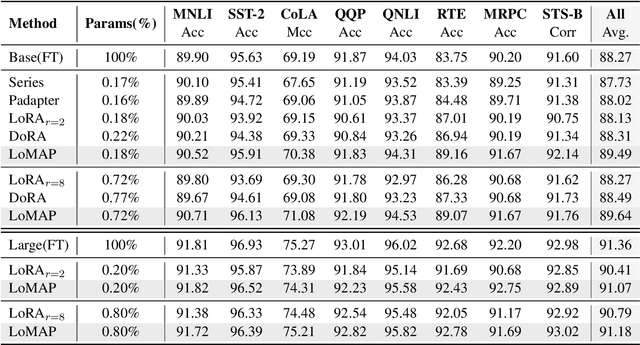

Abstract:The rapid development of large language models has revolutionized natural language processing, but their fine-tuning remains computationally expensive, hindering broad deployment. Parameter-efficient fine-tuning (PEFT) methods, such as LoRA, have emerged as solutions. Recent work like DoRA attempts to further decompose weight adaptation into direction and magnitude components. However, existing formulations often define direction heuristically at the column level, lacking a principled geometric foundation. In this paper, we propose MAP, a novel framework that reformulates weight matrices as high-dimensional vectors and decouples their adaptation into direction and magnitude in a rigorous manner. MAP normalizes the pre-trained weights, learns a directional update, and introduces two scalar coefficients to independently scale the magnitude of the base and update vectors. This design enables more interpretable and flexible adaptation, and can be seamlessly integrated into existing PEFT methods. Extensive experiments show that MAP significantly improves performance when coupling with existing methods, offering a simple yet powerful enhancement to existing PEFT methods. Given the universality and simplicity of MAP, we hope it can serve as a default setting for designing future PEFT methods.
Generalized Tensor-based Parameter-Efficient Fine-Tuning via Lie Group Transformations
Apr 01, 2025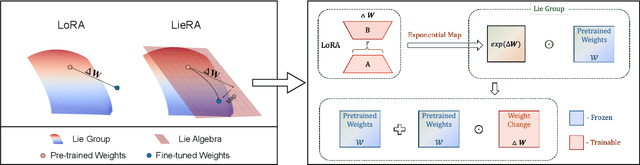
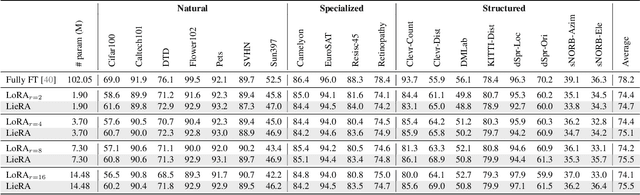
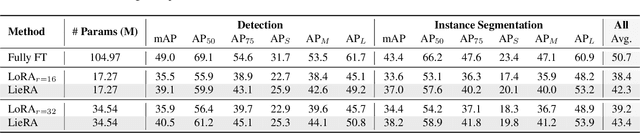

Abstract:Adapting pre-trained foundation models for diverse downstream tasks is a core practice in artificial intelligence. However, the wide range of tasks and high computational costs make full fine-tuning impractical. To overcome this, parameter-efficient fine-tuning (PEFT) methods like LoRA have emerged and are becoming a growing research focus. Despite the success of these methods, they are primarily designed for linear layers, focusing on two-dimensional matrices while largely ignoring higher-dimensional parameter spaces like convolutional kernels. Moreover, directly applying these methods to higher-dimensional parameter spaces often disrupts their structural relationships. Given the rapid advancements in matrix-based PEFT methods, rather than designing a specialized strategy, we propose a generalization that extends matrix-based PEFT methods to higher-dimensional parameter spaces without compromising their structural properties. Specifically, we treat parameters as elements of a Lie group, with updates modeled as perturbations in the corresponding Lie algebra. These perturbations are mapped back to the Lie group through the exponential map, ensuring smooth, consistent updates that preserve the inherent structure of the parameter space. Extensive experiments on computer vision and natural language processing validate the effectiveness and versatility of our approach, demonstrating clear improvements over existing methods.
Multimodal Learning for Embryo Viability Prediction in Clinical IVF
Oct 21, 2024Abstract:In clinical In-Vitro Fertilization (IVF), identifying the most viable embryo for transfer is important to increasing the likelihood of a successful pregnancy. Traditionally, this process involves embryologists manually assessing embryos' static morphological features at specific intervals using light microscopy. This manual evaluation is not only time-intensive and costly, due to the need for expert analysis, but also inherently subjective, leading to variability in the selection process. To address these challenges, we develop a multimodal model that leverages both time-lapse video data and Electronic Health Records (EHRs) to predict embryo viability. One of the primary challenges of our research is to effectively combine time-lapse video and EHR data, owing to their inherent differences in modality. We comprehensively analyze our multimodal model with various modality inputs and integration approaches. Our approach will enable fast and automated embryo viability predictions in scale for clinical IVF.
Unleashing the Power of Task-Specific Directions in Parameter Efficient Fine-tuning
Sep 02, 2024



Abstract:Large language models demonstrate impressive performance on downstream tasks, yet requiring extensive resource consumption when fully fine-tuning all parameters. To mitigate this, Parameter Efficient Fine-Tuning (PEFT) strategies, such as LoRA, have been developed. In this paper, we delve into the concept of task-specific directions--critical for transitioning large models from pre-trained states to task-specific enhancements in PEFT. We propose a framework to clearly define these directions and explore their properties, and practical utilization challenges. We then introduce a novel approach, LoRA-Dash, which aims to maximize the impact of task-specific directions during the fine-tuning process, thereby enhancing model performance on targeted tasks. Extensive experiments have conclusively demonstrated the effectiveness of LoRA-Dash, and in-depth analyses further reveal the underlying mechanisms of LoRA-Dash. The code is available at https://github.com/Chongjie-Si/Subspace-Tuning.
MoRA: LoRA Guided Multi-Modal Disease Diagnosis with Missing Modality
Aug 17, 2024Abstract:Multi-modal pre-trained models efficiently extract and fuse features from different modalities with low memory requirements for fine-tuning. Despite this efficiency, their application in disease diagnosis is under-explored. A significant challenge is the frequent occurrence of missing modalities, which impairs performance. Additionally, fine-tuning the entire pre-trained model demands substantial computational resources. To address these issues, we introduce Modality-aware Low-Rank Adaptation (MoRA), a computationally efficient method. MoRA projects each input to a low intrinsic dimension but uses different modality-aware up-projections for modality-specific adaptation in cases of missing modalities. Practically, MoRA integrates into the first block of the model, significantly improving performance when a modality is missing. It requires minimal computational resources, with less than 1.6% of the trainable parameters needed compared to training the entire model. Experimental results show that MoRA outperforms existing techniques in disease diagnosis, demonstrating superior performance, robustness, and training efficiency.
BeyondCT: A deep learning model for predicting pulmonary function from chest CT scans
Aug 10, 2024Abstract:Abstract Background: Pulmonary function tests (PFTs) and computed tomography (CT) imaging are vital in diagnosing, managing, and monitoring lung diseases. A common issue in practice is the lack of access to recorded pulmonary functions despite available chest CT scans. Purpose: To develop and validate a deep learning algorithm for predicting pulmonary function directly from chest CT scans. Methods: The development cohort came from the Pittsburgh Lung Screening Study (PLuSS) (n=3619). The validation cohort came from the Specialized Centers of Clinically Oriented Research (SCCOR) in COPD (n=662). A deep learning model called BeyondCT, combining a three-dimensional (3D) convolutional neural network (CNN) and Vision Transformer (ViT) architecture, was used to predict forced vital capacity (FVC) and forced expiratory volume in one second (FEV1) from non-contrasted inspiratory chest CT scans. A 3D CNN model without ViT was used for comparison. Subject demographics (age, gender, smoking status) were also incorporated into the model. Performance was compared to actual PFTs using mean absolute error (MAE, L), percentage error, and R square. Results: The 3D-CNN model achieved MAEs of 0.395 L and 0.383 L, percentage errors of 13.84% and 18.85%, and R square of 0.665 and 0.679 for FVC and FEV1, respectively. The BeyondCT model without demographics had MAEs of 0.362 L and 0.371 L, percentage errors of 10.89% and 14.96%, and R square of 0.719 and 0.727, respectively. Including demographics improved performance (p<0.05), with MAEs of 0.356 L and 0.353 L, percentage errors of 10.79% and 14.82%, and R square of 0.77 and 0.739 for FVC and FEV1 in the test set. Conclusion: The BeyondCT model showed robust performance in predicting lung function from non-contrast inspiratory chest CT scans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge