Jiantao Pu
GRN+: A Simplified Generative Reinforcement Network for Tissue Layer Analysis in 3D Ultrasound Images for Chronic Low-back Pain
Mar 25, 2025Abstract:3D ultrasound delivers high-resolution, real-time images of soft tissues, which is essential for pain research. However, manually distinguishing various tissues for quantitative analysis is labor-intensive. To streamline this process, we developed and validated GRN+, a novel multi-model framework that automates layer segmentation with minimal annotated data. GRN+ combines a ResNet-based generator and a U-Net segmentation model. Through a method called Segmentation-guided Enhancement (SGE), the generator produces new images and matching masks under the guidance of the segmentation model, with its weights adjusted according to the segmentation loss gradient. To prevent gradient explosion and secure stable training, a two-stage backpropagation strategy was implemented: the first stage propagates the segmentation loss through both the generator and segmentation model, while the second stage concentrates on optimizing the segmentation model alone, thereby refining mask prediction using the generated images. Tested on 69 fully annotated 3D ultrasound scans from 29 subjects with six manually labeled tissue layers, GRN+ outperformed all other semi-supervised methods in terms of the Dice coefficient using only 5% labeled data, despite not using unlabeled data for unsupervised training. Additionally, when applied to fully annotated datasets, GRN+ with SGE achieved a 2.16% higher Dice coefficient while incurring lower computational costs compared to other models. Overall, GRN+ provides accurate tissue segmentation while reducing both computational expenses and the dependency on extensive annotations, making it an effective tool for 3D ultrasound analysis in cLBP patients.
InterSliceBoost: Identifying Tissue Layers in Three-dimensional Ultrasound Images for Chronic Lower Back Pain (cLBP) Assessment
Mar 25, 2025Abstract:Available studies on chronic lower back pain (cLBP) typically focus on one or a few specific tissues rather than conducting a comprehensive layer-by-layer analysis. Since three-dimensional (3-D) images often contain hundreds of slices, manual annotation of these anatomical structures is both time-consuming and error-prone. We aim to develop and validate a novel approach called InterSliceBoost to enable the training of a segmentation model on a partially annotated dataset without compromising segmentation performance. The architecture of InterSliceBoost includes two components: an inter-slice generator and a segmentation model. The generator utilizes residual block-based encoders to extract features from adjacent image-mask pairs (IMPs). Differential features are calculated and input into a decoder to generate inter-slice IMPs. The segmentation model is trained on partially annotated datasets (e.g., skipping 1, 2, 3, or 7 images) and the generated inter-slice IMPs. To validate the performance of InterSliceBoost, we utilized a dataset of 76 B-mode ultrasound scans acquired on 29 subjects enrolled in an ongoing cLBP study. InterSliceBoost, trained on only 33% of the image slices, achieved a mean Dice coefficient of 80.84% across all six layers on the independent test set, with Dice coefficients of 73.48%, 61.11%, 81.87%, 95.74%, 83.52% and 88.74% for segmenting dermis, superficial fat, superficial fascial membrane, deep fat, deep fascial membrane, and muscle. This performance is significantly higher than the conventional model trained on fully annotated images (p<0.05). InterSliceBoost can effectively segment the six tissue layers depicted on 3-D B-model ultrasound images in settings with partial annotations.
Segmentation-Aware Generative Reinforcement Network (GRN) for Tissue Layer Segmentation in 3-D Ultrasound Images for Chronic Low-back Pain (cLBP) Assessment
Jan 29, 2025Abstract:We introduce a novel segmentation-aware joint training framework called generative reinforcement network (GRN) that integrates segmentation loss feedback to optimize both image generation and segmentation performance in a single stage. An image enhancement technique called segmentation-guided enhancement (SGE) is also developed, where the generator produces images tailored specifically for the segmentation model. Two variants of GRN were also developed, including GRN for sample-efficient learning (GRN-SEL) and GRN for semi-supervised learning (GRN-SSL). GRN's performance was evaluated using a dataset of 69 fully annotated 3D ultrasound scans from 29 subjects. The annotations included six anatomical structures: dermis, superficial fat, superficial fascial membrane (SFM), deep fat, deep fascial membrane (DFM), and muscle. Our results show that GRN-SEL with SGE reduces labeling efforts by up to 70% while achieving a 1.98% improvement in the Dice Similarity Coefficient (DSC) compared to models trained on fully labeled datasets. GRN-SEL alone reduces labeling efforts by 60%, GRN-SSL with SGE decreases labeling requirements by 70%, and GRN-SSL alone by 60%, all while maintaining performance comparable to fully supervised models. These findings suggest the effectiveness of the GRN framework in optimizing segmentation performance with significantly less labeled data, offering a scalable and efficient solution for ultrasound image analysis and reducing the burdens associated with data annotation.
BeyondCT: A deep learning model for predicting pulmonary function from chest CT scans
Aug 10, 2024Abstract:Abstract Background: Pulmonary function tests (PFTs) and computed tomography (CT) imaging are vital in diagnosing, managing, and monitoring lung diseases. A common issue in practice is the lack of access to recorded pulmonary functions despite available chest CT scans. Purpose: To develop and validate a deep learning algorithm for predicting pulmonary function directly from chest CT scans. Methods: The development cohort came from the Pittsburgh Lung Screening Study (PLuSS) (n=3619). The validation cohort came from the Specialized Centers of Clinically Oriented Research (SCCOR) in COPD (n=662). A deep learning model called BeyondCT, combining a three-dimensional (3D) convolutional neural network (CNN) and Vision Transformer (ViT) architecture, was used to predict forced vital capacity (FVC) and forced expiratory volume in one second (FEV1) from non-contrasted inspiratory chest CT scans. A 3D CNN model without ViT was used for comparison. Subject demographics (age, gender, smoking status) were also incorporated into the model. Performance was compared to actual PFTs using mean absolute error (MAE, L), percentage error, and R square. Results: The 3D-CNN model achieved MAEs of 0.395 L and 0.383 L, percentage errors of 13.84% and 18.85%, and R square of 0.665 and 0.679 for FVC and FEV1, respectively. The BeyondCT model without demographics had MAEs of 0.362 L and 0.371 L, percentage errors of 10.89% and 14.96%, and R square of 0.719 and 0.727, respectively. Including demographics improved performance (p<0.05), with MAEs of 0.356 L and 0.353 L, percentage errors of 10.79% and 14.82%, and R square of 0.77 and 0.739 for FVC and FEV1 in the test set. Conclusion: The BeyondCT model showed robust performance in predicting lung function from non-contrast inspiratory chest CT scans.
Assessment of central serous chorioretinopathy (CSC) depicted on color fundus photographs using deep Learning
Jan 14, 2019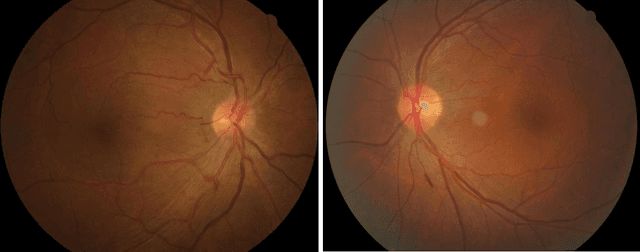

Abstract:To investigate whether and to what extent central serous chorioretinopathy (CSC) depicted on color fundus photographs can be assessed using deep learning technology. We collected a total of 2,504 fundus images acquired on different subjects. We verified the CSC status of these images using their corresponding optical coherence tomography (OCT) images. A total of 1,329 images depicted CSC. These images were preprocessed and normalized. This resulting dataset was randomly split into three parts in the ratio of 8:1:1 respectively for training, validation, and testing purposes. We used the deep learning architecture termed InceptionV3 to train the classifier. We performed nonparametric receiver operating characteristic (ROC) analyses to assess the capability of the developed algorithm to identify CSC. The Kappa coefficient between the two raters was 0.48 (p < 0.001), while the Kappa coefficients between the computer and the two raters were 0.59 (p < 0.001) and 0.33 (p < 0.05).Our experiments showed that the computer algorithm based on deep learning can assess CSC depicted on color fundus photographs in a relatively reliable and consistent way.
Performance assessment of the deep learning technologies in grading glaucoma severity
Oct 31, 2018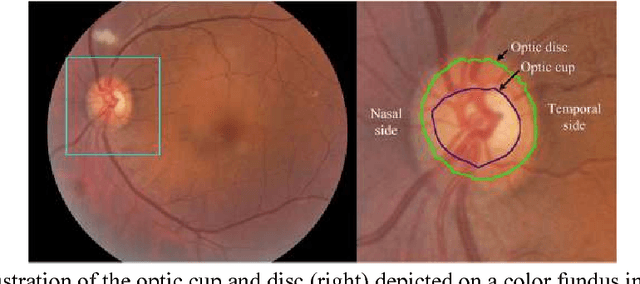
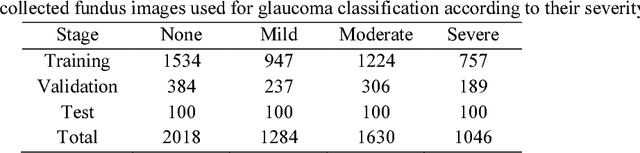
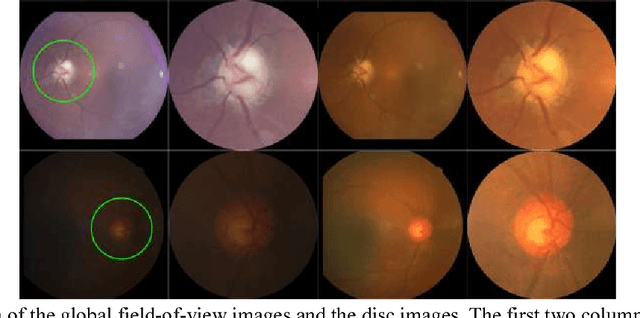
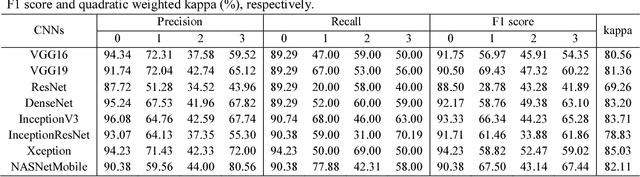
Abstract:Objective: To validate and compare the performance of eight available deep learning architectures in grading the severity of glaucoma based on color fundus images. Materials and Methods: We retrospectively collected a dataset of 5978 fundus images and their glaucoma severities were annotated by the consensus of two experienced ophthalmologists. We preprocessed the images to generate global and local regions of interest (ROIs), namely the global field-of-view images and the local disc region images. We then divided the generated images into three independent sub-groups for training, validation, and testing purposes. With the datasets, eight convolutional neural networks (CNNs) (i.e., VGG16, VGG19, ResNet, DenseNet, InceptionV3, InceptionResNet, Xception, and NASNetMobile) were trained separately to grade glaucoma severity, and validated quantitatively using the area under the receiver operating characteristic (ROC) curve and the quadratic kappa score. Results: The CNNs, except VGG16 and VGG19, achieved average kappa scores of 80.36% and 78.22% when trained from scratch on global and local ROIs, and 85.29% and 82.72% when fine-tuned using the pre-trained weights, respectively. VGG16 and VGG19 achieved reasonable accuracy when trained from scratch, but they failed when using pre-trained weights for global and local ROIs. Among these CNNs, the DenseNet had the highest classification accuracy (i.e., 75.50%) based on pre-trained weights when using global ROIs, as compared to 65.50% when using local ROIs. Conclusion: The experiments demonstrated the feasibility of the deep learning technology in grading glaucoma severity. In particular, global field-of-view images contain relatively richer information that may be critical for glaucoma assessment, suggesting that we should use the entire field-of-view of a fundus image for training a deep learning network.
SDFN: Segmentation-based Deep Fusion Network for Thoracic Disease Classification in Chest X-ray Images
Oct 30, 2018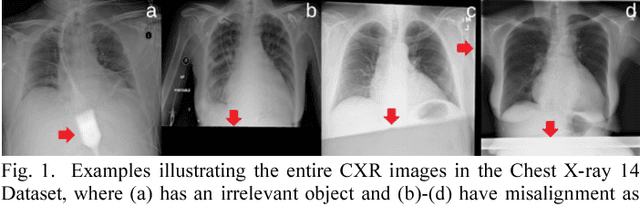
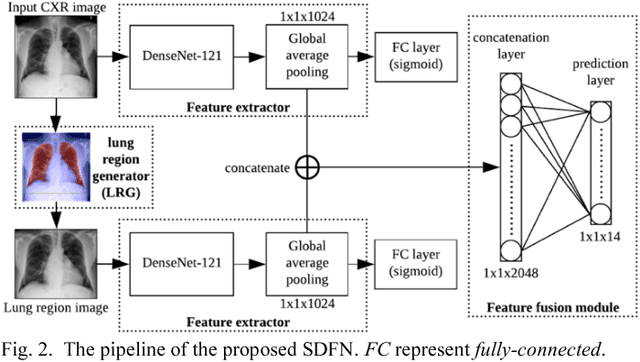
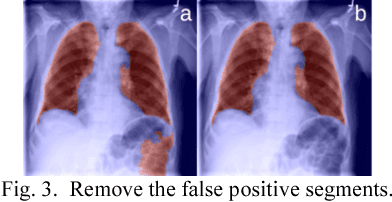
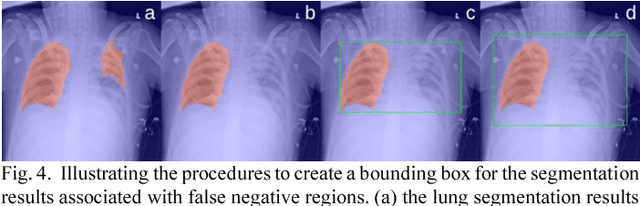
Abstract:This study aims to automatically diagnose thoracic diseases depicted on the chest x-ray (CXR) images using deep convolutional neural networks. The existing methods generally used the entire CXR images for training purposes, but this strategy may suffer from two drawbacks. First, potential misalignment or the existence of irrelevant objects in the entire CXR images may cause unnecessary noise and thus limit the network performance. Second, the relatively low image resolution caused by the resizing operation, which is a common preprocessing procedure for training neural networks, may lead to the loss of image details, making it difficult to detect pathologies with small lesion regions. To address these issues, we present a novel method termed as segmentation-based deep fusion network (SDFN), which leverages the higher-resolution information of local lung regions. Specifically, the local lung regions were identified and cropped by the Lung Region Generator (LRG). Two CNN-based classification models were then used as feature extractors to obtain the discriminative features of the entire CXR images and the cropped lung region images. Lastly, the obtained features were fused by the feature fusion module for disease classification. Evaluated by the NIH benchmark split on the Chest X-ray 14 Dataset, our experimental result demonstrated that the developed method achieved more accurate disease classification compared with the available approaches via the receiver operating characteristic (ROC) analyses. It was also found that the SDFN could localize the lesion regions more precisely as compared to the traditional method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge