Yangkang Zhang
DGL-LifeSci: An Open-Source Toolkit for Deep Learning on Graphs in Life Science
Jun 27, 2021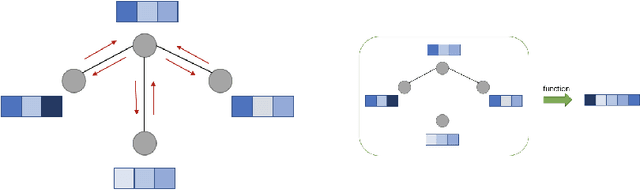
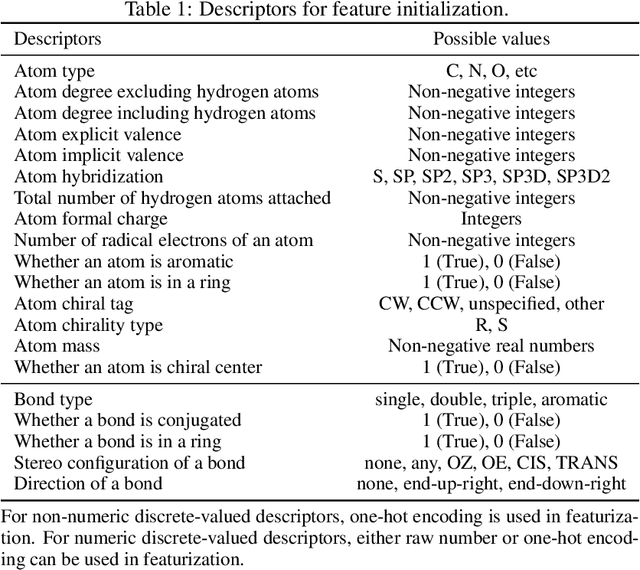
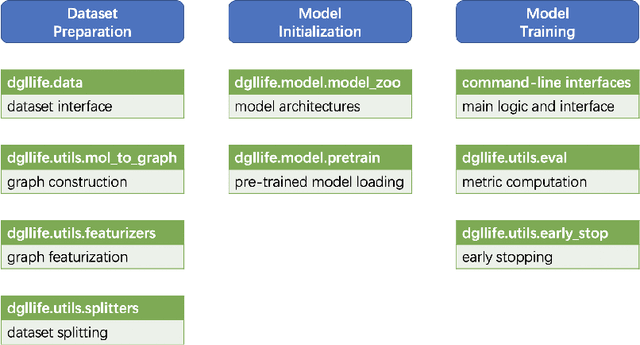

Abstract:Graph neural networks (GNNs) constitute a class of deep learning methods for graph data. They have wide applications in chemistry and biology, such as molecular property prediction, reaction prediction and drug-target interaction prediction. Despite the interest, GNN-based modeling is challenging as it requires graph data pre-processing and modeling in addition to programming and deep learning. Here we present DGL-LifeSci, an open-source package for deep learning on graphs in life science. DGL-LifeSci is a python toolkit based on RDKit, PyTorch and Deep Graph Library (DGL). DGL-LifeSci allows GNN-based modeling on custom datasets for molecular property prediction, reaction prediction and molecule generation. With its command-line interfaces, users can perform modeling without any background in programming and deep learning. We test the command-line interfaces using standard benchmarks MoleculeNet, USPTO, and ZINC. Compared with previous implementations, DGL-LifeSci achieves a speed up by up to 6x. For modeling flexibility, DGL-LifeSci provides well-optimized modules for various stages of the modeling pipeline. In addition, DGL-LifeSci provides pre-trained models for reproducing the test experiment results and applying models without training. The code is distributed under an Apache-2.0 License and is freely accessible at https://github.com/awslabs/dgl-lifesci.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge