Kewei Li
Key Laboratory of Symbolic Computation and Knowledge Engineering of Ministry of Education, Jilin University, Changchun, China, College of Computer Science and Technology, Jilin University, Changchun, China
A transformer-BiGRU-based framework with data augmentation and confident learning for network intrusion detection
Sep 05, 2025



Abstract:In today's fast-paced digital communication, the surge in network traffic data and frequency demands robust and precise network intrusion solutions. Conventional machine learning methods struggle to grapple with complex patterns within the vast network intrusion datasets, which suffer from data scarcity and class imbalance. As a result, we have integrated machine learning and deep learning techniques within the network intrusion detection system to bridge this gap. This study has developed TrailGate, a novel framework that combines machine learning and deep learning techniques. By integrating Transformer and Bidirectional Gated Recurrent Unit (BiGRU) architectures with advanced feature selection strategies and supplemented by data augmentation techniques, TrailGate can identifies common attack types and excels at detecting and mitigating emerging threats. This algorithmic fusion excels at detecting common and well-understood attack types and has the unique ability to swiftly identify and neutralize emerging threats that stem from existing paradigms.
DeepSelective: Feature Gating and Representation Matching for Interpretable Clinical Prediction
Apr 15, 2025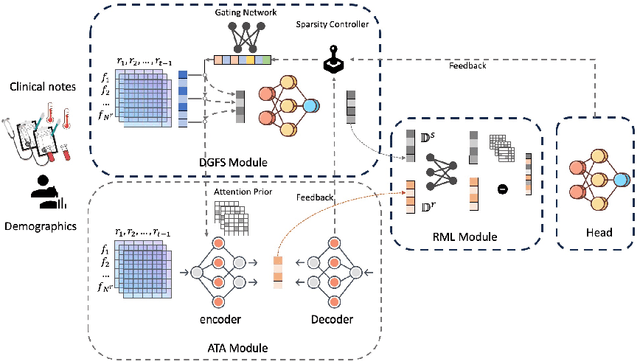
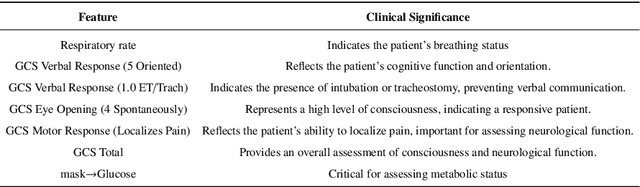
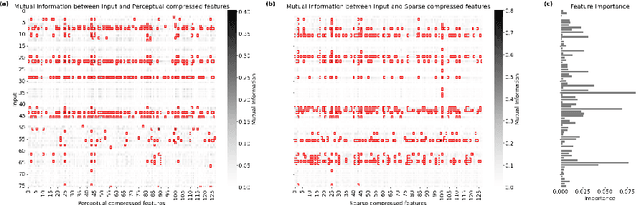
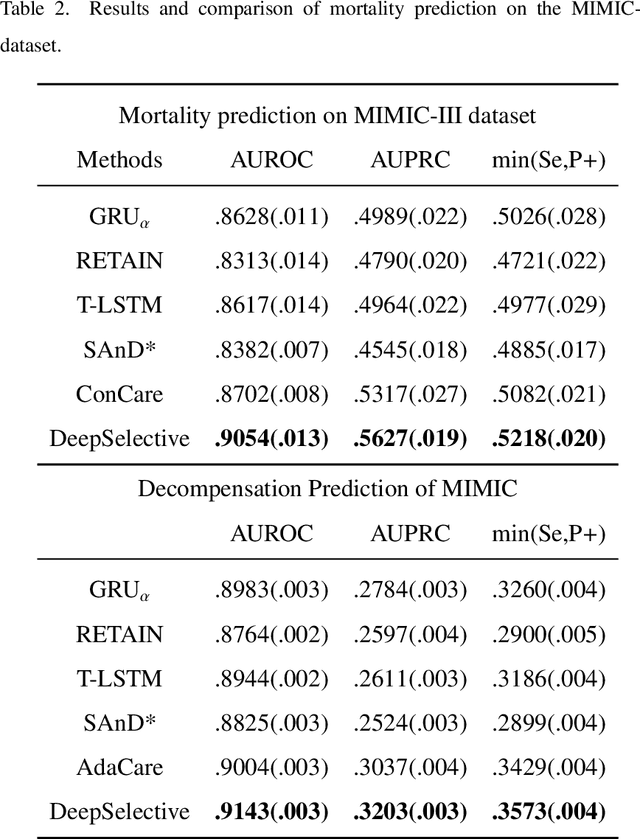
Abstract:The rapid accumulation of Electronic Health Records (EHRs) has transformed healthcare by providing valuable data that enhance clinical predictions and diagnoses. While conventional machine learning models have proven effective, they often lack robust representation learning and depend heavily on expert-crafted features. Although deep learning offers powerful solutions, it is often criticized for its lack of interpretability. To address these challenges, we propose DeepSelective, a novel end to end deep learning framework for predicting patient prognosis using EHR data, with a strong emphasis on enhancing model interpretability. DeepSelective combines data compression techniques with an innovative feature selection approach, integrating custom-designed modules that work together to improve both accuracy and interpretability. Our experiments demonstrate that DeepSelective not only enhances predictive accuracy but also significantly improves interpretability, making it a valuable tool for clinical decision-making. The source code is freely available at http://www.healthinformaticslab.org/supp/resources.php .
Latent Swap Joint Diffusion for Long-Form Audio Generation
Feb 07, 2025



Abstract:Previous work on long-form audio generation using global-view diffusion or iterative generation demands significant training or inference costs. While recent advancements in multi-view joint diffusion for panoramic generation provide an efficient option, they struggle with spectrum generation with severe overlap distortions and high cross-view consistency costs. We initially explore this phenomenon through the connectivity inheritance of latent maps and uncover that averaging operations excessively smooth the high-frequency components of the latent map. To address these issues, we propose Swap Forward (SaFa), a frame-level latent swap framework that synchronizes multiple diffusions to produce a globally coherent long audio with more spectrum details in a forward-only manner. At its core, the bidirectional Self-Loop Latent Swap is applied between adjacent views, leveraging stepwise diffusion trajectory to adaptively enhance high-frequency components without disrupting low-frequency components. Furthermore, to ensure cross-view consistency, the unidirectional Reference-Guided Latent Swap is applied between the reference and the non-overlap regions of each subview during the early stages, providing centralized trajectory guidance. Quantitative and qualitative experiments demonstrate that SaFa significantly outperforms existing joint diffusion methods and even training-based long audio generation models. Moreover, we find that it also adapts well to panoramic generation, achieving comparable state-of-the-art performance with greater efficiency and model generalizability. Project page is available at https://swapforward.github.io/.
Information Entropy Invariance: Enhancing Length Extrapolation in Attention Mechanisms
Jan 15, 2025Abstract:Improving the length extrapolation capabilities of Large Language Models (LLMs) remains a critical challenge in natural language processing. Many recent efforts have focused on modifying the scaled dot-product attention mechanism, and often introduce scaled temperatures without rigorous theoretical justification. To fill this gap, we introduce a novel approach based on information entropy invariance. We propose two new scaled temperatures to enhance length extrapolation. First, a training-free method InfoScale is designed for dot-product attention, and preserves focus on original tokens during length extrapolation by ensuring information entropy remains consistent. Second, we theoretically analyze the impact of scaling (CosScale) on cosine attention. Experimental data demonstrates that combining InfoScale and CosScale achieves state-of-the-art performance on the GAU-{\alpha} model with a context window extended to 64 times the training length, and outperforms seven existing methods. Our analysis reveals that significantly increasing CosScale approximates windowed attention, and highlights the significance of attention score dilution as a key challenge in long-range context handling. The code and data are available at https://github.com/HT-NEKO/InfoScale.
Enhancing Multimodal Sentiment Analysis for Missing Modality through Self-Distillation and Unified Modality Cross-Attention
Oct 19, 2024


Abstract:In multimodal sentiment analysis, collecting text data is often more challenging than video or audio due to higher annotation costs and inconsistent automatic speech recognition (ASR) quality. To address this challenge, our study has developed a robust model that effectively integrates multimodal sentiment information, even in the absence of text modality. Specifically, we have developed a Double-Flow Self-Distillation Framework, including Unified Modality Cross-Attention (UMCA) and Modality Imagination Autoencoder (MIA), which excels at processing both scenarios with complete modalities and those with missing text modality. In detail, when the text modality is missing, our framework uses the LLM-based model to simulate the text representation from the audio modality, while the MIA module supplements information from the other two modalities to make the simulated text representation similar to the real text representation. To further align the simulated and real representations, and to enable the model to capture the continuous nature of sample orders in sentiment valence regression tasks, we have also introduced the Rank-N Contrast (RNC) loss function. When testing on the CMU-MOSEI, our model achieved outstanding performance on MAE and significantly outperformed other models when text modality is missing. The code is available at: https://github.com/WarmCongee/SDUMC
TemporalPaD: a reinforcement-learning framework for temporal feature representation and dimension reduction
Sep 27, 2024



Abstract:Recent advancements in feature representation and dimension reduction have highlighted their crucial role in enhancing the efficacy of predictive modeling. This work introduces TemporalPaD, a novel end-to-end deep learning framework designed for temporal pattern datasets. TemporalPaD integrates reinforcement learning (RL) with neural networks to achieve concurrent feature representation and feature reduction. The framework consists of three cooperative modules: a Policy Module, a Representation Module, and a Classification Module, structured based on the Actor-Critic (AC) framework. The Policy Module, responsible for dimensionality reduction through RL, functions as the actor, while the Representation Module for feature extraction and the Classification Module collectively serve as the critic. We comprehensively evaluate TemporalPaD using 29 UCI datasets, a well-known benchmark for validating feature reduction algorithms, through 10 independent tests and 10-fold cross-validation. Additionally, given that TemporalPaD is specifically designed for time series data, we apply it to a real-world DNA classification problem involving enhancer category and enhancer strength. The results demonstrate that TemporalPaD is an efficient and effective framework for achieving feature reduction, applicable to both structured data and sequence datasets. The source code of the proposed TemporalPaD is freely available as supplementary material to this article and at http://www.healthinformaticslab.org/supp/.
Towards Automated Data Sciences with Natural Language and SageCopilot: Practices and Lessons Learned
Jul 21, 2024



Abstract:While the field of NL2SQL has made significant advancements in translating natural language instructions into executable SQL scripts for data querying and processing, achieving full automation within the broader data science pipeline - encompassing data querying, analysis, visualization, and reporting - remains a complex challenge. This study introduces SageCopilot, an advanced, industry-grade system system that automates the data science pipeline by integrating Large Language Models (LLMs), Autonomous Agents (AutoAgents), and Language User Interfaces (LUIs). Specifically, SageCopilot incorporates a two-phase design: an online component refining users' inputs into executable scripts through In-Context Learning (ICL) and running the scripts for results reporting & visualization, and an offline preparing demonstrations requested by ICL in the online phase. A list of trending strategies such as Chain-of-Thought and prompt-tuning have been used to augment SageCopilot for enhanced performance. Through rigorous testing and comparative analysis against prompt-based solutions, SageCopilot has been empirically validated to achieve superior end-to-end performance in generating or executing scripts and offering results with visualization, backed by real-world datasets. Our in-depth ablation studies highlight the individual contributions of various components and strategies used by SageCopilot to the end-to-end correctness for data sciences.
AMPCliff: quantitative definition and benchmarking of activity cliffs in antimicrobial peptides
Apr 15, 2024



Abstract:Activity cliff (AC) is a phenomenon that a pair of similar molecules differ by a small structural alternation but exhibit a large difference in their biochemical activities. The AC of small molecules has been extensively investigated but limited knowledge is accumulated about the AC phenomenon in peptides with canonical amino acids. This study introduces a quantitative definition and benchmarking framework AMPCliff for the AC phenomenon in antimicrobial peptides (AMPs) composed by canonical amino acids. A comprehensive analysis of the existing AMP dataset reveals a significant prevalence of AC within AMPs. AMPCliff quantifies the activities of AMPs by the metric minimum inhibitory concentration (MIC), and defines 0.9 as the minimum threshold for the normalized BLOSUM62 similarity score between a pair of aligned peptides with at least two-fold MIC changes. This study establishes a benchmark dataset of paired AMPs in Staphylococcus aureus from the publicly available AMP dataset GRAMPA, and conducts a rigorous procedure to evaluate various AMP AC prediction models, including nine machine learning, four deep learning algorithms, four masked language models, and four generative language models. Our analysis reveals that these models are capable of detecting AMP AC events and the pre-trained protein language ESM2 model demonstrates superior performance across the evaluations. The predictive performance of AMP activity cliffs remains to be further improved, considering that ESM2 with 33 layers only achieves the Spearman correlation coefficient=0.50 for the regression task of the MIC values on the benchmark dataset. Source code and additional resources are available at https://www.healthinformaticslab.org/supp/ or https://github.com/Kewei2023/AMPCliff-generation.
PepHarmony: A Multi-View Contrastive Learning Framework for Integrated Sequence and Structure-Based Peptide Encoding
Jan 21, 2024



Abstract:Recent advances in protein language models have catalyzed significant progress in peptide sequence representation. Despite extensive exploration in this field, pre-trained models tailored for peptide-specific needs remain largely unaddressed due to the difficulty in capturing the complex and sometimes unstable structures of peptides. This study introduces a novel multi-view contrastive learning framework PepHarmony for the sequence-based peptide encoding task. PepHarmony innovatively combines both sequence- and structure-level information into a sequence-level encoding module through contrastive learning. We carefully select datasets from the Protein Data Bank (PDB) and AlphaFold database to encompass a broad spectrum of peptide sequences and structures. The experimental data highlights PepHarmony's exceptional capability in capturing the intricate relationship between peptide sequences and structures compared with the baseline and fine-tuned models. The robustness of our model is confirmed through extensive ablation studies, which emphasize the crucial roles of contrastive loss and strategic data sorting in enhancing predictive performance. The proposed PepHarmony framework serves as a notable contribution to peptide representations, and offers valuable insights for future applications in peptide drug discovery and peptide engineering. We have made all the source code utilized in this study publicly accessible via GitHub at https://github.com/zhangruochi/PepHarmony or http://www.healthinformaticslab.org/supp/.
PepLand: a large-scale pre-trained peptide representation model for a comprehensive landscape of both canonical and non-canonical amino acids
Nov 08, 2023Abstract:In recent years, the scientific community has become increasingly interested on peptides with non-canonical amino acids due to their superior stability and resistance to proteolytic degradation. These peptides present promising modifications to biological, pharmacological, and physiochemical attributes in both endogenous and engineered peptides. Notwithstanding their considerable advantages, the scientific community exhibits a conspicuous absence of an effective pre-trained model adept at distilling feature representations from such complex peptide sequences. We herein propose PepLand, a novel pre-training architecture for representation and property analysis of peptides spanning both canonical and non-canonical amino acids. In essence, PepLand leverages a comprehensive multi-view heterogeneous graph neural network tailored to unveil the subtle structural representations of peptides. Empirical validations underscore PepLand's effectiveness across an array of peptide property predictions, encompassing protein-protein interactions, permeability, solubility, and synthesizability. The rigorous evaluation confirms PepLand's unparalleled capability in capturing salient synthetic peptide features, thereby laying a robust foundation for transformative advances in peptide-centric research domains. We have made all the source code utilized in this study publicly accessible via GitHub at https://github.com/zhangruochi/pepland
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge