Caihua Shan
VidGuard-R1: AI-Generated Video Detection and Explanation via Reasoning MLLMs and RL
Oct 02, 2025Abstract:With the rapid advancement of AI-generated videos, there is an urgent need for effective detection tools to mitigate societal risks such as misinformation and reputational harm. In addition to accurate classification, it is essential that detection models provide interpretable explanations to ensure transparency for regulators and end users. To address these challenges, we introduce VidGuard-R1, the first video authenticity detector that fine-tunes a multi-modal large language model (MLLM) using group relative policy optimization (GRPO). Our model delivers both highly accurate judgments and insightful reasoning. We curate a challenging dataset of 140k real and AI-generated videos produced by state-of-the-art generation models, carefully designing the generation process to maximize discrimination difficulty. We then fine-tune Qwen-VL using GRPO with two specialized reward models that target temporal artifacts and generation complexity. Extensive experiments demonstrate that VidGuard-R1 achieves state-of-the-art zero-shot performance on existing benchmarks, with additional training pushing accuracy above 95%. Case studies further show that VidGuard-R1 produces precise and interpretable rationales behind its predictions. The code is publicly available at https://VidGuard-R1.github.io.
Chain-of-Model Learning for Language Model
May 17, 2025Abstract:In this paper, we propose a novel learning paradigm, termed Chain-of-Model (CoM), which incorporates the causal relationship into the hidden states of each layer as a chain style, thereby introducing great scaling efficiency in model training and inference flexibility in deployment. We introduce the concept of Chain-of-Representation (CoR), which formulates the hidden states at each layer as a combination of multiple sub-representations (i.e., chains) at the hidden dimension level. In each layer, each chain from the output representations can only view all of its preceding chains in the input representations. Consequently, the model built upon CoM framework can progressively scale up the model size by increasing the chains based on the previous models (i.e., chains), and offer multiple sub-models at varying sizes for elastic inference by using different chain numbers. Based on this principle, we devise Chain-of-Language-Model (CoLM), which incorporates the idea of CoM into each layer of Transformer architecture. Based on CoLM, we further introduce CoLM-Air by introducing a KV sharing mechanism, that computes all keys and values within the first chain and then shares across all chains. This design demonstrates additional extensibility, such as enabling seamless LM switching, prefilling acceleration and so on. Experimental results demonstrate our CoLM family can achieve comparable performance to the standard Transformer, while simultaneously enabling greater flexiblity, such as progressive scaling to improve training efficiency and offer multiple varying model sizes for elastic inference, paving a a new way toward building language models. Our code will be released in the future at: https://github.com/microsoft/CoLM.
Omni-DNA: A Unified Genomic Foundation Model for Cross-Modal and Multi-Task Learning
Feb 05, 2025



Abstract:Large Language Models (LLMs) demonstrate remarkable generalizability across diverse tasks, yet genomic foundation models (GFMs) still require separate finetuning for each downstream application, creating significant overhead as model sizes grow. Moreover, existing GFMs are constrained by rigid output formats, limiting their applicability to various genomic tasks. In this work, we revisit the transformer-based auto-regressive models and introduce Omni-DNA, a family of cross-modal multi-task models ranging from 20 million to 1 billion parameters. Our approach consists of two stages: (i) pretraining on DNA sequences with next token prediction objective, and (ii) expanding the multi-modal task-specific tokens and finetuning for multiple downstream tasks simultaneously. When evaluated on the Nucleotide Transformer and GB benchmarks, Omni-DNA achieves state-of-the-art performance on 18 out of 26 tasks. Through multi-task finetuning, Omni-DNA addresses 10 acetylation and methylation tasks at once, surpassing models trained on each task individually. Finally, we design two complex genomic tasks, DNA2Function and Needle-in-DNA, which map DNA sequences to textual functional descriptions and images, respectively, indicating Omni-DNA's cross-modal capabilities to broaden the scope of genomic applications. All the models are available through https://huggingface.co/collections/zehui127
A Comprehensive Analysis on LLM-based Node Classification Algorithms
Feb 02, 2025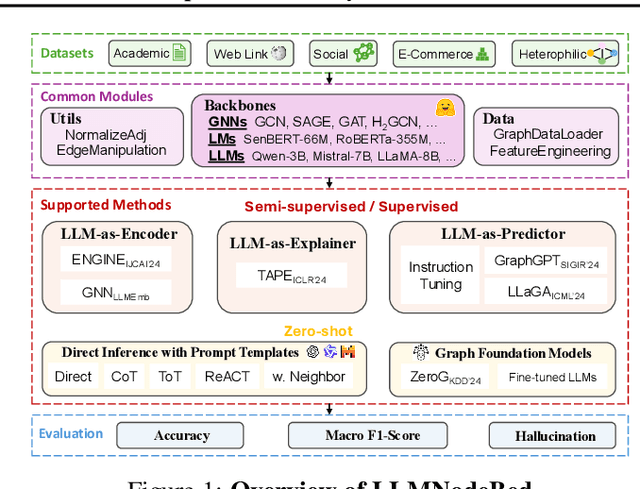
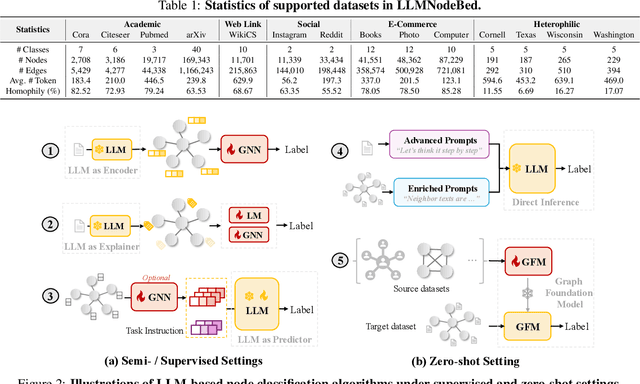
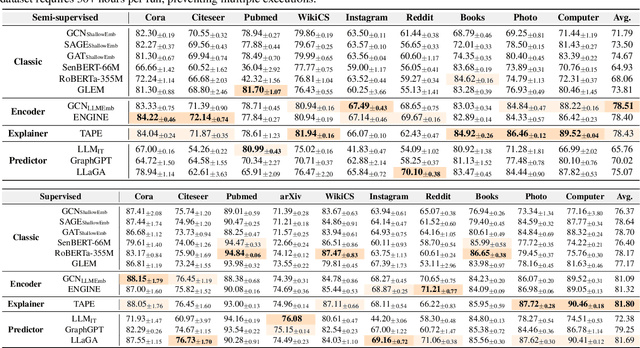
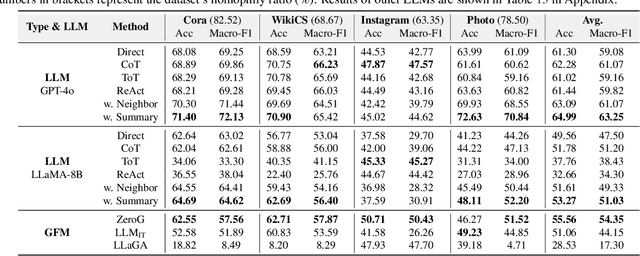
Abstract:Node classification is a fundamental task in graph analysis, with broad applications across various fields. Recent breakthroughs in Large Language Models (LLMs) have enabled LLM-based approaches for this task. Although many studies demonstrate the impressive performance of LLM-based methods, the lack of clear design guidelines may hinder their practical application. In this work, we aim to establish such guidelines through a fair and systematic comparison of these algorithms. As a first step, we developed LLMNodeBed, a comprehensive codebase and testbed for node classification using LLMs. It includes ten datasets, eight LLM-based algorithms, and three learning paradigms, and is designed for easy extension with new methods and datasets. Subsequently, we conducted extensive experiments, training and evaluating over 2,200 models, to determine the key settings (e.g., learning paradigms and homophily) and components (e.g., model size) that affect performance. Our findings uncover eight insights, e.g., (1) LLM-based methods can significantly outperform traditional methods in a semi-supervised setting, while the advantage is marginal in a supervised setting; (2) Graph Foundation Models can beat open-source LLMs but still fall short of strong LLMs like GPT-4o in a zero-shot setting. We hope that the release of LLMNodeBed, along with our insights, will facilitate reproducible research and inspire future studies in this field. Codes and datasets are released at \href{https://llmnodebed.github.io/}{https://llmnodebed.github.io/}.
How Do Large Language Models Understand Graph Patterns? A Benchmark for Graph Pattern Comprehension
Oct 04, 2024



Abstract:Benchmarking the capabilities and limitations of large language models (LLMs) in graph-related tasks is becoming an increasingly popular and crucial area of research. Recent studies have shown that LLMs exhibit a preliminary ability to understand graph structures and node features. However, the potential of LLMs in graph pattern mining remains largely unexplored. This is a key component in fields such as computational chemistry, biology, and social network analysis. To bridge this gap, this work introduces a comprehensive benchmark to assess LLMs' capabilities in graph pattern tasks. We have developed a benchmark that evaluates whether LLMs can understand graph patterns based on either terminological or topological descriptions. Additionally, our benchmark tests the LLMs' capacity to autonomously discover graph patterns from data. The benchmark encompasses both synthetic and real datasets, and a variety of models, with a total of 11 tasks and 7 models. Our experimental framework is designed for easy expansion to accommodate new models and datasets. Our findings reveal that: (1) LLMs have preliminary abilities to understand graph patterns, with O1-mini outperforming in the majority of tasks; (2) Formatting input data to align with the knowledge acquired during pretraining can enhance performance; (3) The strategies employed by LLMs may differ from those used in conventional algorithms.
Revisiting the Graph Reasoning Ability of Large Language Models: Case Studies in Translation, Connectivity and Shortest Path
Aug 18, 2024
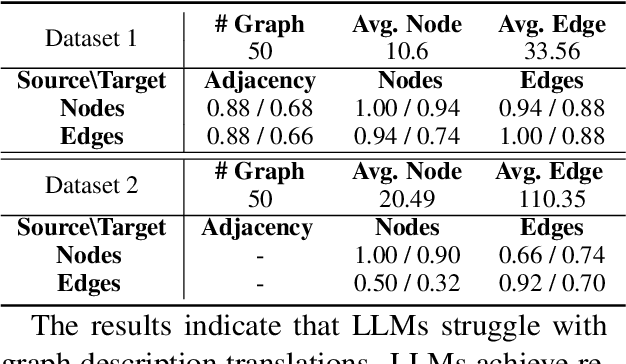

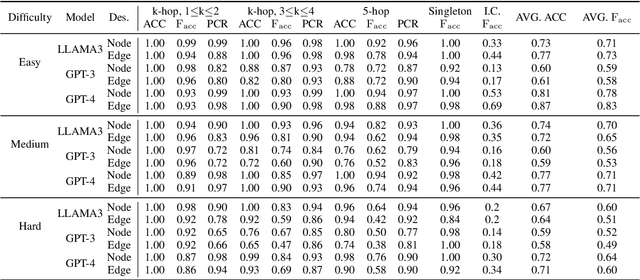
Abstract:Large Language Models (LLMs) have achieved great success in various reasoning tasks. In this work, we focus on the graph reasoning ability of LLMs. Although theoretical studies proved that LLMs are capable of handling graph reasoning tasks, empirical evaluations reveal numerous failures. To deepen our understanding on this discrepancy, we revisit the ability of LLMs on three fundamental graph tasks: graph description translation, graph connectivity, and the shortest-path problem. Our findings suggest that LLMs can fail to understand graph structures through text descriptions and exhibit varying performance for all these three fundamental tasks. Meanwhile, we perform a real-world investigation on knowledge graphs and make consistent observations with our findings. The codes and datasets are available.
Can Graph Learning Improve Task Planning?
May 29, 2024Abstract:Task planning is emerging as an important research topic alongside the development of large language models (LLMs). It aims to break down complex user requests into solvable sub-tasks, thereby fulfilling the original requests. In this context, the sub-tasks can be naturally viewed as a graph, where the nodes represent the sub-tasks, and the edges denote the dependencies among them. Consequently, task planning is a decision-making problem that involves selecting a connected path or subgraph within the corresponding graph and invoking it. In this paper, we explore graph learning-based methods for task planning, a direction that is orthogonal to the prevalent focus on prompt design. Our interest in graph learning stems from a theoretical discovery: the biases of attention and auto-regressive loss impede LLMs' ability to effectively navigate decision-making on graphs, which is adeptly addressed by graph neural networks (GNNs). This theoretical insight led us to integrate GNNs with LLMs to enhance overall performance. Extensive experiments demonstrate that GNN-based methods surpass existing solutions even without training, and minimal training can further enhance their performance. Additionally, our approach complements prompt engineering and fine-tuning techniques, with performance further enhanced by improved prompts or a fine-tuned model.
Learning from Graphs with Heterophily: Progress and Future
Feb 01, 2024Abstract:Graphs are structured data that models complex relations between real-world entities. Heterophilous graphs, where linked nodes are prone to be with different labels or dissimilar features, have recently attracted significant attention and found many applications. Meanwhile, increasing efforts have been made to advance learning from heterophilous graphs. Although there exist surveys on the relevant topic, they focus on heterophilous GNNs, which are only sub-topics of heterophilous graph learning. In this survey, we comprehensively overview existing works on learning from graphs with heterophily.First, we collect over 180 publications and introduce the development of this field. Then, we systematically categorize existing methods based on a hierarchical taxonomy including learning strategies, model architectures and practical applications. Finally, we discuss the primary challenges of existing studies and highlight promising avenues for future research.More publication details and corresponding open-source codes can be accessed and will be continuously updated at our repositories:https://github.com/gongchenghua/Awesome-Survey-Graphs-with-Heterophily.
AdaMedGraph: Adaboosting Graph Neural Networks for Personalized Medicine
Nov 24, 2023Abstract:Precision medicine tailored to individual patients has gained significant attention in recent times. Machine learning techniques are now employed to process personalized data from various sources, including images, genetics, and assessments. These techniques have demonstrated good outcomes in many clinical prediction tasks. Notably, the approach of constructing graphs by linking similar patients and then applying graph neural networks (GNNs) stands out, because related information from analogous patients are aggregated and considered for prediction. However, selecting the appropriate edge feature to define patient similarity and construct the graph is challenging, given that each patient is depicted by high-dimensional features from diverse sources. Previous studies rely on human expertise to select the edge feature, which is neither scalable nor efficient in pinpointing crucial edge features for complex diseases. In this paper, we propose a novel algorithm named \ours, which can automatically select important features to construct multiple patient similarity graphs, and train GNNs based on these graphs as weak learners in adaptive boosting. \ours{} is evaluated on two real-world medical scenarios and shows superiors performance.
Prioritized Propagation in Graph Neural Networks
Nov 06, 2023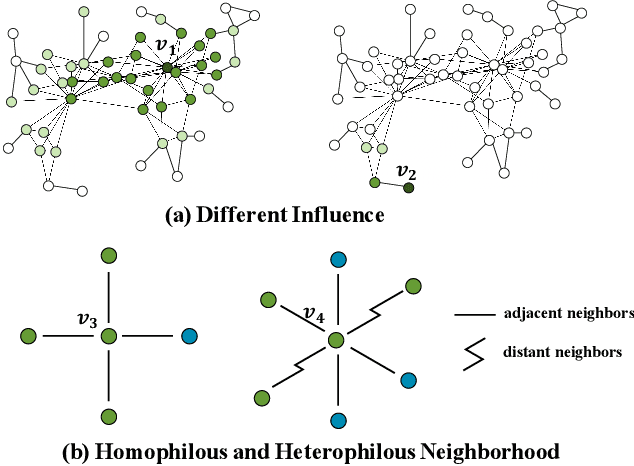
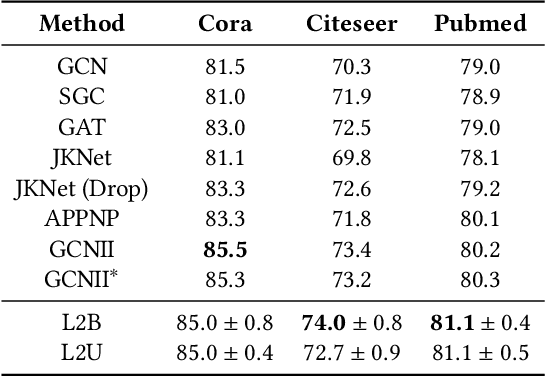
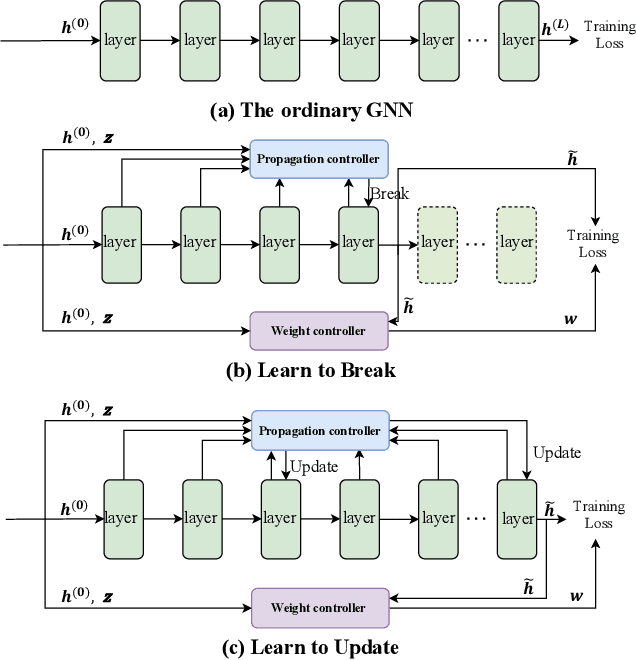
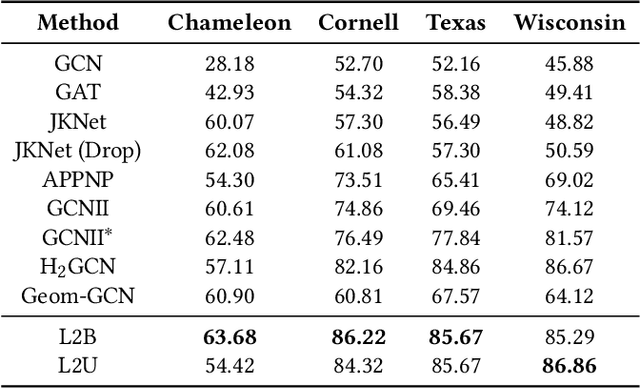
Abstract:Graph neural networks (GNNs) have recently received significant attention. Learning node-wise message propagation in GNNs aims to set personalized propagation steps for different nodes in the graph. Despite the success, existing methods ignore node priority that can be reflected by node influence and heterophily. In this paper, we propose a versatile framework PPro, which can be integrated with most existing GNN models and aim to learn prioritized node-wise message propagation in GNNs. Specifically, the framework consists of three components: a backbone GNN model, a propagation controller to determine the optimal propagation steps for nodes, and a weight controller to compute the priority scores for nodes. We design a mutually enhanced mechanism to compute node priority, optimal propagation step and label prediction. We also propose an alternative optimization strategy to learn the parameters in the backbone GNN model and two parametric controllers. We conduct extensive experiments to compare our framework with other 11 state-of-the-art competitors on 8 benchmark datasets. Experimental results show that our framework can lead to superior performance in terms of propagation strategies and node representations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge