Xinnan Dai
Exploring Graph Tasks with Pure LLMs: A Comprehensive Benchmark and Investigation
Feb 26, 2025Abstract:Graph-structured data has become increasingly prevalent across various domains, raising the demand for effective models to handle graph tasks like node classification and link prediction. Traditional graph learning models like Graph Neural Networks (GNNs) have made significant strides, but their capabilities in handling graph data remain limited in certain contexts. In recent years, large language models (LLMs) have emerged as promising candidates for graph tasks, yet most studies focus primarily on performance benchmarks and fail to address their broader potential, including their ability to handle limited data, their transferability across tasks, and their robustness. In this work, we provide a comprehensive exploration of LLMs applied to graph tasks. We evaluate the performance of pure LLMs, including those without parameter optimization and those fine-tuned with instructions, across various scenarios. Our analysis goes beyond accuracy, assessing LLM ability to perform in few-shot/zero-shot settings, transfer across domains, understand graph structures, and demonstrate robustness in challenging scenarios. We conduct extensive experiments with 16 graph learning models alongside 6 LLMs (e.g., Llama3B, GPT-4o, Qwen-plus), comparing their performance on datasets like Cora, PubMed, ArXiv, and Products. Our findings show that LLMs, particularly those with instruction tuning, outperform traditional models in few-shot settings, exhibit strong domain transferability, and demonstrate excellent generalization and robustness. This work offers valuable insights into the capabilities of LLMs for graph learning, highlighting their advantages and potential for real-world applications, and paving the way for future research in this area. Codes and datasets are released in https://github.com/myflashbarry/LLM-benchmarking.
How Do Large Language Models Understand Graph Patterns? A Benchmark for Graph Pattern Comprehension
Oct 04, 2024



Abstract:Benchmarking the capabilities and limitations of large language models (LLMs) in graph-related tasks is becoming an increasingly popular and crucial area of research. Recent studies have shown that LLMs exhibit a preliminary ability to understand graph structures and node features. However, the potential of LLMs in graph pattern mining remains largely unexplored. This is a key component in fields such as computational chemistry, biology, and social network analysis. To bridge this gap, this work introduces a comprehensive benchmark to assess LLMs' capabilities in graph pattern tasks. We have developed a benchmark that evaluates whether LLMs can understand graph patterns based on either terminological or topological descriptions. Additionally, our benchmark tests the LLMs' capacity to autonomously discover graph patterns from data. The benchmark encompasses both synthetic and real datasets, and a variety of models, with a total of 11 tasks and 7 models. Our experimental framework is designed for easy expansion to accommodate new models and datasets. Our findings reveal that: (1) LLMs have preliminary abilities to understand graph patterns, with O1-mini outperforming in the majority of tasks; (2) Formatting input data to align with the knowledge acquired during pretraining can enhance performance; (3) The strategies employed by LLMs may differ from those used in conventional algorithms.
Revisiting the Graph Reasoning Ability of Large Language Models: Case Studies in Translation, Connectivity and Shortest Path
Aug 18, 2024
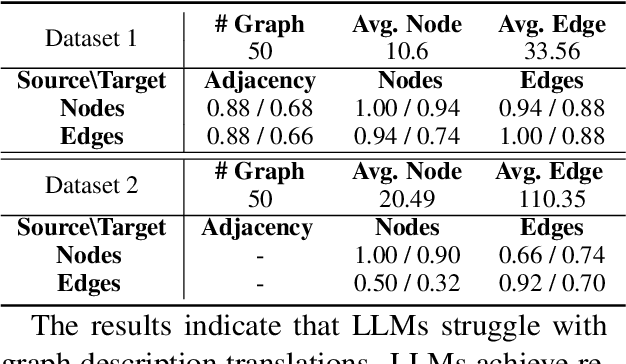

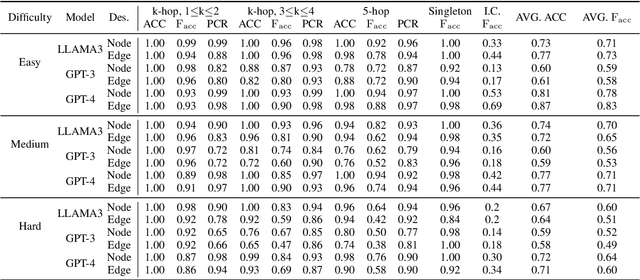
Abstract:Large Language Models (LLMs) have achieved great success in various reasoning tasks. In this work, we focus on the graph reasoning ability of LLMs. Although theoretical studies proved that LLMs are capable of handling graph reasoning tasks, empirical evaluations reveal numerous failures. To deepen our understanding on this discrepancy, we revisit the ability of LLMs on three fundamental graph tasks: graph description translation, graph connectivity, and the shortest-path problem. Our findings suggest that LLMs can fail to understand graph structures through text descriptions and exhibit varying performance for all these three fundamental tasks. Meanwhile, we perform a real-world investigation on knowledge graphs and make consistent observations with our findings. The codes and datasets are available.
Biological Factor Regulatory Neural Network
Apr 11, 2023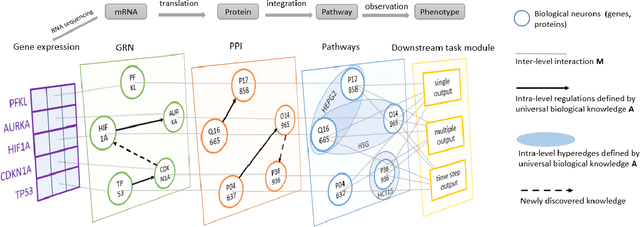


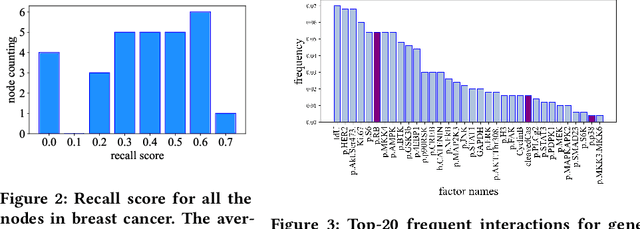
Abstract:Genes are fundamental for analyzing biological systems and many recent works proposed to utilize gene expression for various biological tasks by deep learning models. Despite their promising performance, it is hard for deep neural networks to provide biological insights for humans due to their black-box nature. Recently, some works integrated biological knowledge with neural networks to improve the transparency and performance of their models. However, these methods can only incorporate partial biological knowledge, leading to suboptimal performance. In this paper, we propose the Biological Factor Regulatory Neural Network (BFReg-NN), a generic framework to model relations among biological factors in cell systems. BFReg-NN starts from gene expression data and is capable of merging most existing biological knowledge into the model, including the regulatory relations among genes or proteins (e.g., gene regulatory networks (GRN), protein-protein interaction networks (PPI)) and the hierarchical relations among genes, proteins and pathways (e.g., several genes/proteins are contained in a pathway). Moreover, BFReg-NN also has the ability to provide new biologically meaningful insights because of its white-box characteristics. Experimental results on different gene expression-based tasks verify the superiority of BFReg-NN compared with baselines. Our case studies also show that the key insights found by BFReg-NN are consistent with the biological literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge