Multiple Instance Learning
Multiple instance learning is a machine learning paradigm where training data is organized into bags of instances.
Papers and Code
Interpretable Rheumatoid Arthritis Scoring via Anatomy-aware Multiple Instance Learning
Aug 08, 2025The Sharp/van der Heijde (SvdH) score has been widely used in clinical trials to quantify radiographic damage in Rheumatoid Arthritis (RA), but its complexity has limited its adoption in routine clinical practice. To address the inefficiency of manual scoring, this work proposes a two-stage pipeline for interpretable image-level SvdH score prediction using dual-hand radiographs. Our approach extracts disease-relevant image regions and integrates them using attention-based multiple instance learning to generate image-level features for prediction. We propose two region extraction schemes: 1) sampling image tiles most likely to contain abnormalities, and 2) cropping patches containing disease-relevant joints. With Scheme 2, our best individual score prediction model achieved a Pearson's correlation coefficient (PCC) of 0.943 and a root mean squared error (RMSE) of 15.73. Ensemble learning further boosted prediction accuracy, yielding a PCC of 0.945 and RMSE of 15.57, achieving state-of-the-art performance that is comparable to that of experienced radiologists (PCC = 0.97, RMSE = 18.75). Finally, our pipeline effectively identified and made decisions based on anatomical structures which clinicians consider relevant to RA progression.
Priority-Aware Pathological Hierarchy Training for Multiple Instance Learning
Jul 28, 2025Multiple Instance Learning (MIL) is increasingly being used as a support tool within clinical settings for pathological diagnosis decisions, achieving high performance and removing the annotation burden. However, existing approaches for clinical MIL tasks have not adequately addressed the priority issues that exist in relation to pathological symptoms and diagnostic classes, causing MIL models to ignore priority among classes. To overcome this clinical limitation of MIL, we propose a new method that addresses priority issues using two hierarchies: vertical inter-hierarchy and horizontal intra-hierarchy. The proposed method aligns MIL predictions across each hierarchical level and employs an implicit feature re-usability during training to facilitate clinically more serious classes within the same level. Experiments with real-world patient data show that the proposed method effectively reduces misdiagnosis and prioritizes more important symptoms in multiclass scenarios. Further analysis verifies the efficacy of the proposed components and qualitatively confirms the MIL predictions against challenging cases with multiple symptoms.
Backdoor Vectors: a Task Arithmetic View on Backdoor Attacks and Defenses
Oct 09, 2025Model merging (MM) recently emerged as an effective method for combining large deep learning models. However, it poses significant security risks. Recent research shows that it is highly susceptible to backdoor attacks, which introduce a hidden trigger into a single fine-tuned model instance that allows the adversary to control the output of the final merged model at inference time. In this work, we propose a simple framework for understanding backdoor attacks by treating the attack itself as a task vector. $Backdoor\ Vector\ (BV)$ is calculated as the difference between the weights of a fine-tuned backdoored model and fine-tuned clean model. BVs reveal new insights into attacks understanding and a more effective framework to measure their similarity and transferability. Furthermore, we propose a novel method that enhances backdoor resilience through merging dubbed $Sparse\ Backdoor\ Vector\ (SBV)$ that combines multiple attacks into a single one. We identify the core vulnerability behind backdoor threats in MM: $inherent\ triggers$ that exploit adversarial weaknesses in the base model. To counter this, we propose $Injection\ BV\ Subtraction\ (IBVS)$ - an assumption-free defense against backdoors in MM. Our results show that SBVs surpass prior attacks and is the first method to leverage merging to improve backdoor effectiveness. At the same time, IBVS provides a lightweight, general defense that remains effective even when the backdoor threat is entirely unknown.
ERGO: Efficient High-Resolution Visual Understanding for Vision-Language Models
Sep 26, 2025Efficient processing of high-resolution images is crucial for real-world vision-language applications. However, existing Large Vision-Language Models (LVLMs) incur substantial computational overhead due to the large number of vision tokens. With the advent of "thinking with images" models, reasoning now extends beyond text to the visual domain. This capability motivates our two-stage "coarse-to-fine" reasoning pipeline: first, a downsampled image is analyzed to identify task-relevant regions; then, only these regions are cropped at full resolution and processed in a subsequent reasoning stage. This approach reduces computational cost while preserving fine-grained visual details where necessary. A major challenge lies in inferring which regions are truly relevant to a given query. Recent related methods often fail in the first stage after input-image downsampling, due to perception-driven reasoning, where clear visual information is required for effective reasoning. To address this issue, we propose ERGO (Efficient Reasoning & Guided Observation) that performs reasoning-driven perception-leveraging multimodal context to determine where to focus. Our model can account for perceptual uncertainty, expanding the cropped region to cover visually ambiguous areas for answering questions. To this end, we develop simple yet effective reward components in a reinforcement learning framework for coarse-to-fine perception. Across multiple datasets, our approach delivers higher accuracy than the original model and competitive methods, with greater efficiency. For instance, ERGO surpasses Qwen2.5-VL-7B on the V* benchmark by 4.7 points while using only 23% of the vision tokens, achieving a 3x inference speedup. The code and models can be found at: https://github.com/nota-github/ERGO.
Controllable Latent Space Augmentation for Digital Pathology
Aug 20, 2025Whole slide image (WSI) analysis in digital pathology presents unique challenges due to the gigapixel resolution of WSIs and the scarcity of dense supervision signals. While Multiple Instance Learning (MIL) is a natural fit for slide-level tasks, training robust models requires large and diverse datasets. Even though image augmentation techniques could be utilized to increase data variability and reduce overfitting, implementing them effectively is not a trivial task. Traditional patch-level augmentation is prohibitively expensive due to the large number of patches extracted from each WSI, and existing feature-level augmentation methods lack control over transformation semantics. We introduce HistAug, a fast and efficient generative model for controllable augmentations in the latent space for digital pathology. By conditioning on explicit patch-level transformations (e.g., hue, erosion), HistAug generates realistic augmented embeddings while preserving initial semantic information. Our method allows the processing of a large number of patches in a single forward pass efficiently, while at the same time consistently improving MIL model performance. Experiments across multiple slide-level tasks and diverse organs show that HistAug outperforms existing methods, particularly in low-data regimes. Ablation studies confirm the benefits of learned transformations over noise-based perturbations and highlight the importance of uniform WSI-wise augmentation. Code is available at https://github.com/MICS-Lab/HistAug.
IPGPhormer: Interpretable Pathology Graph-Transformer for Survival Analysis
Aug 17, 2025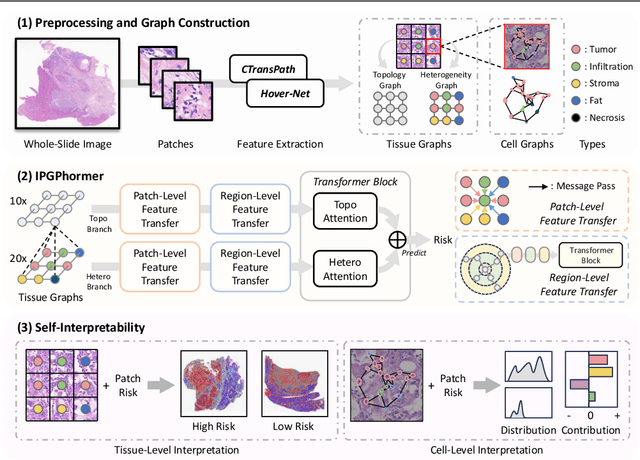
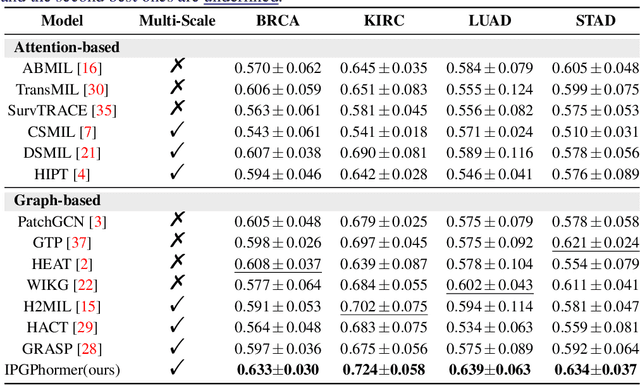
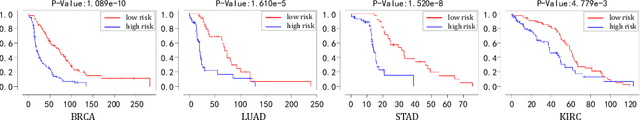
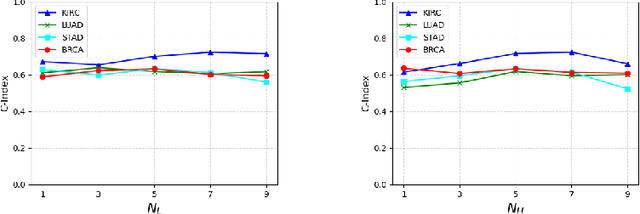
Pathological images play an essential role in cancer prognosis, while survival analysis, which integrates computational techniques, can predict critical clinical events such as patient mortality or disease recurrence from whole-slide images (WSIs). Recent advancements in multiple instance learning have significantly improved the efficiency of survival analysis. However, existing methods often struggle to balance the modeling of long-range spatial relationships with local contextual dependencies and typically lack inherent interpretability, limiting their clinical utility. To address these challenges, we propose the Interpretable Pathology Graph-Transformer (IPGPhormer), a novel framework that captures the characteristics of the tumor microenvironment and models their spatial dependencies across the tissue. IPGPhormer uniquely provides interpretability at both tissue and cellular levels without requiring post-hoc manual annotations, enabling detailed analyses of individual WSIs and cross-cohort assessments. Comprehensive evaluations on four public benchmark datasets demonstrate that IPGPhormer outperforms state-of-the-art methods in both predictive accuracy and interpretability. In summary, our method, IPGPhormer, offers a promising tool for cancer prognosis assessment, paving the way for more reliable and interpretable decision-support systems in pathology. The code is publicly available at https://anonymous.4open.science/r/IPGPhormer-6EEB.
Conditional Representation Learning for Customized Tasks
Oct 06, 2025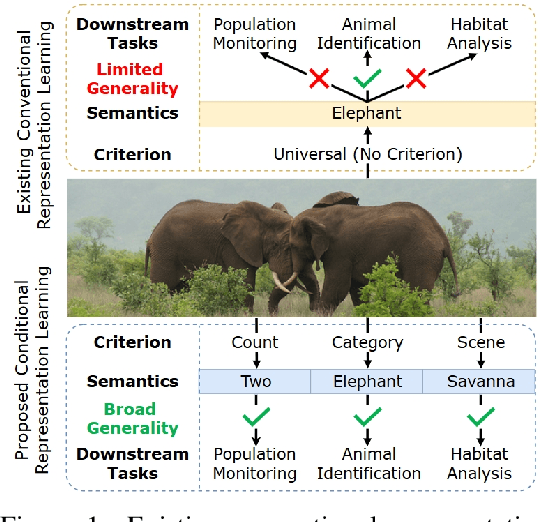

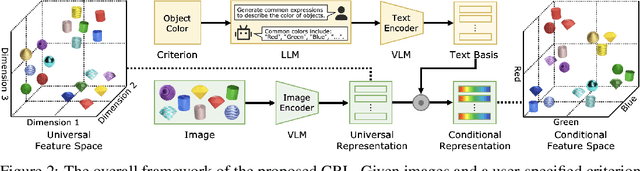
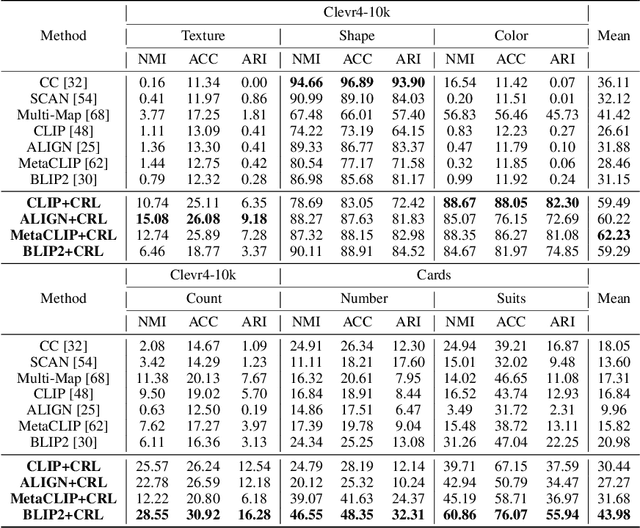
Conventional representation learning methods learn a universal representation that primarily captures dominant semantics, which may not always align with customized downstream tasks. For instance, in animal habitat analysis, researchers prioritize scene-related features, whereas universal embeddings emphasize categorical semantics, leading to suboptimal results. As a solution, existing approaches resort to supervised fine-tuning, which however incurs high computational and annotation costs. In this paper, we propose Conditional Representation Learning (CRL), aiming to extract representations tailored to arbitrary user-specified criteria. Specifically, we reveal that the semantics of a space are determined by its basis, thereby enabling a set of descriptive words to approximate the basis for a customized feature space. Building upon this insight, given a user-specified criterion, CRL first employs a large language model (LLM) to generate descriptive texts to construct the semantic basis, then projects the image representation into this conditional feature space leveraging a vision-language model (VLM). The conditional representation better captures semantics for the specific criterion, which could be utilized for multiple customized tasks. Extensive experiments on classification and retrieval tasks demonstrate the superiority and generality of the proposed CRL. The code is available at https://github.com/XLearning-SCU/2025-NeurIPS-CRL.
Live(r) Die: Predicting Survival in Colorectal Liver Metastasis
Sep 10, 2025Colorectal cancer frequently metastasizes to the liver, significantly reducing long-term survival. While surgical resection is the only potentially curative treatment for colorectal liver metastasis (CRLM), patient outcomes vary widely depending on tumor characteristics along with clinical and genomic factors. Current prognostic models, often based on limited clinical or molecular features, lack sufficient predictive power, especially in multifocal CRLM cases. We present a fully automated framework for surgical outcome prediction from pre- and post-contrast MRI acquired before surgery. Our framework consists of a segmentation pipeline and a radiomics pipeline. The segmentation pipeline learns to segment the liver, tumors, and spleen from partially annotated data by leveraging promptable foundation models to complete missing labels. Also, we propose SAMONAI, a novel zero-shot 3D prompt propagation algorithm that leverages the Segment Anything Model to segment 3D regions of interest from a single point prompt, significantly improving our segmentation pipeline's accuracy and efficiency. The predicted pre- and post-contrast segmentations are then fed into our radiomics pipeline, which extracts features from each tumor and predicts survival using SurvAMINN, a novel autoencoder-based multiple instance neural network for survival analysis. SurvAMINN jointly learns dimensionality reduction and hazard prediction from right-censored survival data, focusing on the most aggressive tumors. Extensive evaluation on an institutional dataset comprising 227 patients demonstrates that our framework surpasses existing clinical and genomic biomarkers, delivering a C-index improvement exceeding 10%. Our results demonstrate the potential of integrating automated segmentation algorithms and radiomics-based survival analysis to deliver accurate, annotation-efficient, and interpretable outcome prediction in CRLM.
Instance-Optimal Matrix Multiplicative Weight Update and Its Quantum Applications
Sep 10, 2025The Matrix Multiplicative Weight Update (MMWU) is a seminal online learning algorithm with numerous applications. Applied to the matrix version of the Learning from Expert Advice (LEA) problem on the $d$-dimensional spectraplex, it is well known that MMWU achieves the minimax-optimal regret bound of $O(\sqrt{T\log d})$, where $T$ is the time horizon. In this paper, we present an improved algorithm achieving the instance-optimal regret bound of $O(\sqrt{T\cdot S(X||d^{-1}I_d)})$, where $X$ is the comparator in the regret, $I_d$ is the identity matrix, and $S(\cdot||\cdot)$ denotes the quantum relative entropy. Furthermore, our algorithm has the same computational complexity as MMWU, indicating that the improvement in the regret bound is ``free''. Technically, we first develop a general potential-based framework for matrix LEA, with MMWU being its special case induced by the standard exponential potential. Then, the crux of our analysis is a new ``one-sided'' Jensen's trace inequality built on a Laplace transform technique, which allows the application of general potential functions beyond exponential to matrix LEA. Our algorithm is finally induced by an optimal potential function from the vector LEA problem, based on the imaginary error function. Complementing the above, we provide a memory lower bound for matrix LEA, and explore the applications of our algorithm in quantum learning theory. We show that it outperforms the state of the art for learning quantum states corrupted by depolarization noise, random quantum states, and Gibbs states. In addition, applying our algorithm to linearized convex losses enables predicting nonlinear quantum properties, such as purity, quantum virtual cooling, and R\'{e}nyi-$2$ correlation.
MOC: Meta-Optimized Classifier for Few-Shot Whole Slide Image Classification
Aug 13, 2025Recent advances in histopathology vision-language foundation models (VLFMs) have shown promise in addressing data scarcity for whole slide image (WSI) classification via zero-shot adaptation. However, these methods remain outperformed by conventional multiple instance learning (MIL) approaches trained on large datasets, motivating recent efforts to enhance VLFM-based WSI classification through fewshot learning paradigms. While existing few-shot methods improve diagnostic accuracy with limited annotations, their reliance on conventional classifier designs introduces critical vulnerabilities to data scarcity. To address this problem, we propose a Meta-Optimized Classifier (MOC) comprising two core components: (1) a meta-learner that automatically optimizes a classifier configuration from a mixture of candidate classifiers and (2) a classifier bank housing diverse candidate classifiers to enable a holistic pathological interpretation. Extensive experiments demonstrate that MOC outperforms prior arts in multiple few-shot benchmarks. Notably, on the TCGA-NSCLC benchmark, MOC improves AUC by 10.4% over the state-of-the-art few-shot VLFM-based methods, with gains up to 26.25% under 1-shot conditions, offering a critical advancement for clinical deployments where diagnostic training data is severely limited. Code is available at https://github.com/xmed-lab/MOC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge