Jin Yang
U-Harmony: Enhancing Joint Training for Segmentation Models with Universal Harmonization
Jan 21, 2026Abstract:In clinical practice, medical segmentation datasets are often limited and heterogeneous, with variations in modalities, protocols, and anatomical targets across institutions. Existing deep learning models struggle to jointly learn from such diverse data, often sacrificing either generalization or domain-specific knowledge. To overcome these challenges, we propose a joint training method called Universal Harmonization (U-Harmony), which can be integrated into deep learning-based architectures with a domain-gated head, enabling a single segmentation model to learn from heterogeneous datasets simultaneously. By integrating U-Harmony, our approach sequentially normalizes and then denormalizes feature distributions to mitigate domain-specific variations while preserving original dataset-specific knowledge. More appealingly, our framework also supports universal modality adaptation, allowing the seamless learning of new imaging modalities and anatomical classes. Extensive experiments on cross-institutional brain lesion datasets demonstrate the effectiveness of our approach, establishing a new benchmark for robust and adaptable 3D medical image segmentation models in real-world clinical settings.
Embodied Robot Manipulation in the Era of Foundation Models: Planning and Learning Perspectives
Dec 28, 2025Abstract:Recent advances in vision, language, and multimodal learning have substantially accelerated progress in robotic foundation models, with robot manipulation remaining a central and challenging problem. This survey examines robot manipulation from an algorithmic perspective and organizes recent learning-based approaches within a unified abstraction of high-level planning and low-level control. At the high level, we extend the classical notion of task planning to include reasoning over language, code, motion, affordances, and 3D representations, emphasizing their role in structured and long-horizon decision making. At the low level, we propose a training-paradigm-oriented taxonomy for learning-based control, organizing existing methods along input modeling, latent representation learning, and policy learning. Finally, we identify open challenges and prospective research directions related to scalability, data efficiency, multimodal physical interaction, and safety. Together, these analyses aim to clarify the design space of modern foundation models for robotic manipulation.
Every Step Evolves: Scaling Reinforcement Learning for Trillion-Scale Thinking Model
Oct 21, 2025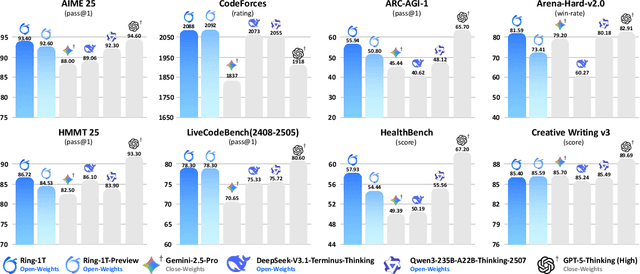
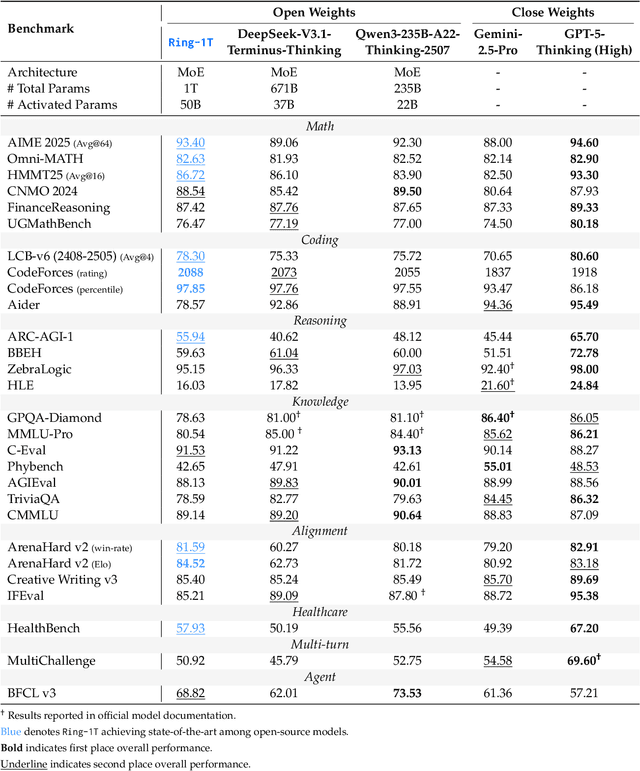
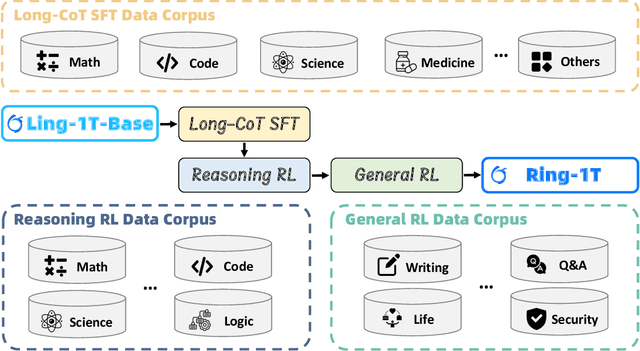
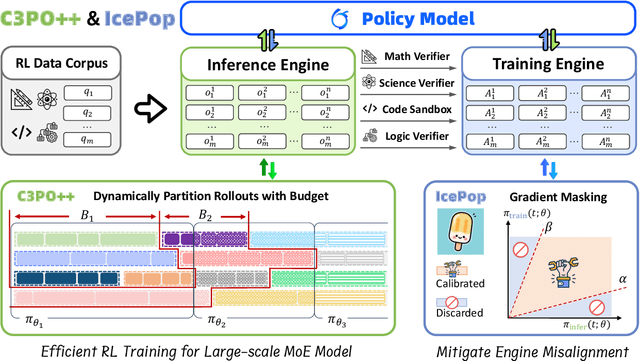
Abstract:We present Ring-1T, the first open-source, state-of-the-art thinking model with a trillion-scale parameter. It features 1 trillion total parameters and activates approximately 50 billion per token. Training such models at a trillion-parameter scale introduces unprecedented challenges, including train-inference misalignment, inefficiencies in rollout processing, and bottlenecks in the RL system. To address these, we pioneer three interconnected innovations: (1) IcePop stabilizes RL training via token-level discrepancy masking and clipping, resolving instability from training-inference mismatches; (2) C3PO++ improves resource utilization for long rollouts under a token budget by dynamically partitioning them, thereby obtaining high time efficiency; and (3) ASystem, a high-performance RL framework designed to overcome the systemic bottlenecks that impede trillion-parameter model training. Ring-1T delivers breakthrough results across critical benchmarks: 93.4 on AIME-2025, 86.72 on HMMT-2025, 2088 on CodeForces, and 55.94 on ARC-AGI-v1. Notably, it attains a silver medal-level result on the IMO-2025, underscoring its exceptional reasoning capabilities. By releasing the complete 1T parameter MoE model to the community, we provide the research community with direct access to cutting-edge reasoning capabilities. This contribution marks a significant milestone in democratizing large-scale reasoning intelligence and establishes a new baseline for open-source model performance.
Beyond Benchmark: LLMs Evaluation with an Anthropomorphic and Value-oriented Roadmap
Aug 26, 2025Abstract:For Large Language Models (LLMs), a disconnect persists between benchmark performance and real-world utility. Current evaluation frameworks remain fragmented, prioritizing technical metrics while neglecting holistic assessment for deployment. This survey introduces an anthropomorphic evaluation paradigm through the lens of human intelligence, proposing a novel three-dimensional taxonomy: Intelligence Quotient (IQ)-General Intelligence for foundational capacity, Emotional Quotient (EQ)-Alignment Ability for value-based interactions, and Professional Quotient (PQ)-Professional Expertise for specialized proficiency. For practical value, we pioneer a Value-oriented Evaluation (VQ) framework assessing economic viability, social impact, ethical alignment, and environmental sustainability. Our modular architecture integrates six components with an implementation roadmap. Through analysis of 200+ benchmarks, we identify key challenges including dynamic assessment needs and interpretability gaps. It provides actionable guidance for developing LLMs that are technically proficient, contextually relevant, and ethically sound. We maintain a curated repository of open-source evaluation resources at: https://github.com/onejune2018/Awesome-LLM-Eval.
Beyond Scaling Law: A Data-Efficient Distillation Framework for Reasoning
Aug 13, 2025Abstract:Large language models (LLMs) demonstrate remarkable reasoning capabilities in tasks such as algorithmic coding and mathematical problem-solving. Recent methods have improved reasoning through expanded corpus and multistage training combining reinforcement learning and supervised fine-tuning. Although some methods suggest that small but targeted dataset can incentivize reasoning via only distillation, a reasoning scaling laws is still taking shape, increasing computational costs. To address this, we propose a data-efficient distillation framework (DED) that optimizes the Pareto frontier of reasoning distillation. Inspired by the on-policy learning and diverse roll-out strategies of reinforcement learning, the key idea of our approach is threefold: (1) We identify that benchmark scores alone do not determine an effective teacher model. Through comprehensive comparisons of leading reasoning LLMs, we develop a method to select an optimal teacher model. (2) While scaling distillation can enhance reasoning, it often degrades out-of-domain performance. A carefully curated, smaller corpus achieves a balanced trade-off between in-domain and out-of-domain capabilities. (3) Diverse reasoning trajectories encourage the student model to develop robust reasoning skills. We validate our method through evaluations on mathematical reasoning (AIME 2024/2025, MATH-500) and code generation (LiveCodeBench), achieving state-of-the-art results with only 0.8k carefully curated examples, bypassing the need for extensive scaling. Our systematic analysis demonstrates that DED outperforms existing methods by considering factors beyond superficial hardness, token length, or teacher model capability. This work offers a practical and efficient pathway to advanced reasoning while preserving general capabilities.
Quality-of-Service Aware LLM Routing for Edge Computing with Multiple Experts
Aug 01, 2025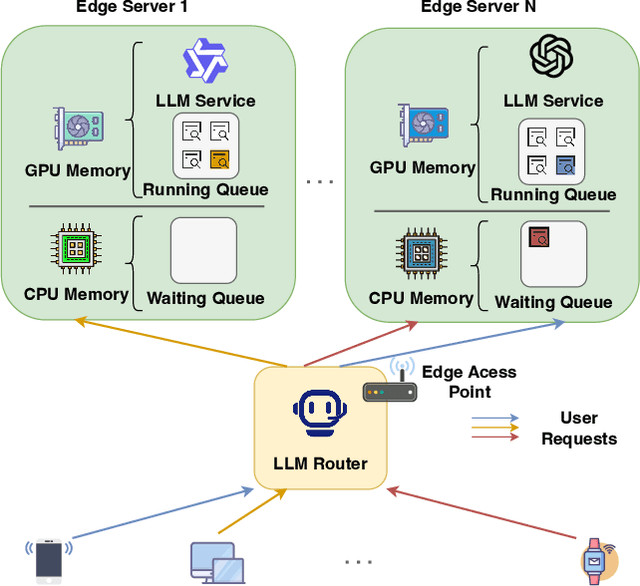
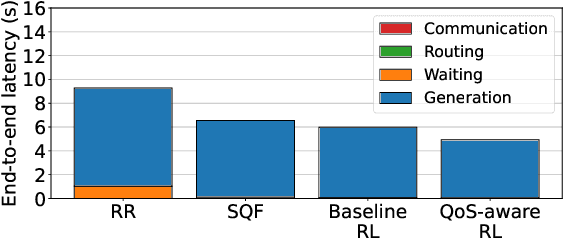
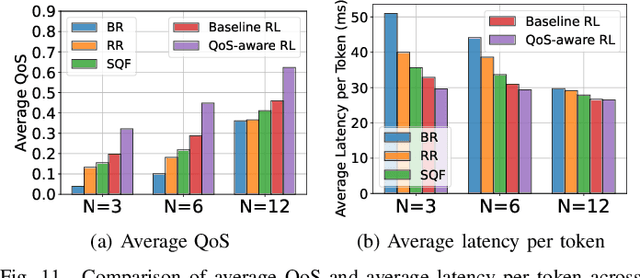
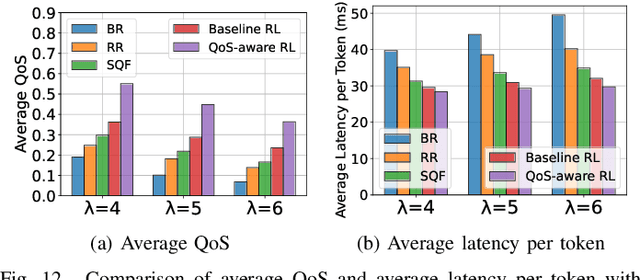
Abstract:Large Language Models (LLMs) have demonstrated remarkable capabilities, leading to a significant increase in user demand for LLM services. However, cloud-based LLM services often suffer from high latency, unstable responsiveness, and privacy concerns. Therefore, multiple LLMs are usually deployed at the network edge to boost real-time responsiveness and protect data privacy, particularly for many emerging smart mobile and IoT applications. Given the varying response quality and latency of LLM services, a critical issue is how to route user requests from mobile and IoT devices to an appropriate LLM service (i.e., edge LLM expert) to ensure acceptable quality-of-service (QoS). Existing routing algorithms fail to simultaneously address the heterogeneity of LLM services, the interference among requests, and the dynamic workloads necessary for maintaining long-term stable QoS. To meet these challenges, in this paper we propose a novel deep reinforcement learning (DRL)-based QoS-aware LLM routing framework for sustained high-quality LLM services. Due to the dynamic nature of the global state, we propose a dynamic state abstraction technique to compactly represent global state features with a heterogeneous graph attention network (HAN). Additionally, we introduce an action impact estimator and a tailored reward function to guide the DRL agent in maximizing QoS and preventing latency violations. Extensive experiments on both Poisson and real-world workloads demonstrate that our proposed algorithm significantly improves average QoS and computing resource efficiency compared to existing baselines.
Step-Audio 2 Technical Report
Jul 24, 2025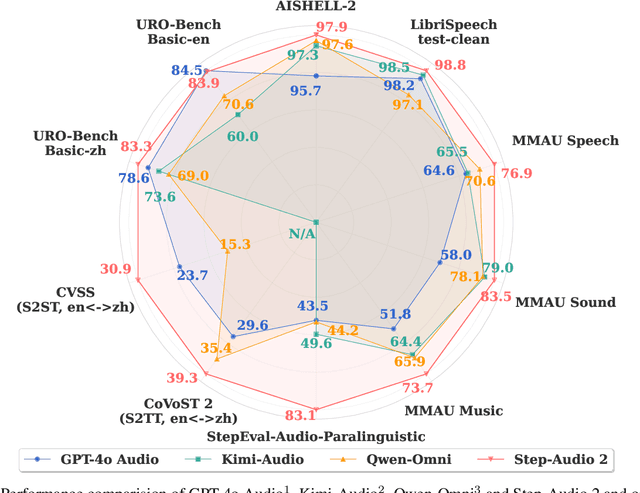

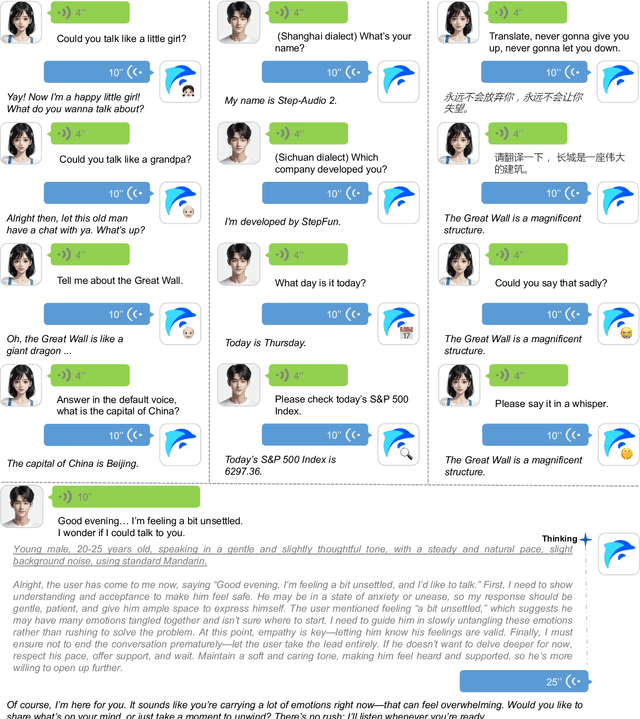

Abstract:This paper presents Step-Audio 2, an end-to-end multi-modal large language model designed for industry-strength audio understanding and speech conversation. By integrating a latent audio encoder and reasoning-centric reinforcement learning (RL), Step-Audio 2 achieves promising performance in automatic speech recognition (ASR) and audio understanding. To facilitate genuine end-to-end speech conversation, Step-Audio 2 incorporates the generation of discrete audio tokens into language modeling, significantly enhancing its responsiveness to paralinguistic information such as speaking styles and emotions. To effectively leverage the rich textual and acoustic knowledge in real-world data, Step-Audio 2 integrates retrieval-augmented generation (RAG) and is able to call external tools such as web search to mitigate hallucination and audio search to switch timbres. Trained on millions of hours of speech and audio data, Step-Audio 2 delivers intelligence and expressiveness across diverse conversational scenarios. Evaluation results demonstrate that Step-Audio 2 achieves state-of-the-art performance on various audio understanding and conversational benchmarks compared to other open-source and commercial solutions. Please visit https://github.com/stepfun-ai/Step-Audio2 for more information.
Bubble Dynamics Transformer: Microrheology at Ultra-High Strain Rates
Jun 13, 2025Abstract:Laser-induced inertial cavitation (LIC)-where microscale vapor bubbles nucleate due to a focused high-energy pulsed laser and then violently collapse under surrounding high local pressures-offers a unique opportunity to investigate soft biological material mechanics at extremely high strain rates (>1000 1/s). Traditional rheological tools are often limited in these regimes by loading speed, resolution, or invasiveness. Here we introduce novel machine learning (ML) based microrheological frameworks that leverage LIC to characterize the viscoelastic properties of biological materials at ultra-high strain rates. We utilize ultra-high-speed imaging to capture time-resolved bubble radius dynamics during LIC events in various soft viscoelastic materials. These bubble radius versus time measurements are then analyzed using a newly developed Bubble Dynamics Transformer (BDT), a neural network trained on physics-based simulation data. The BDT accurately infers material viscoelastic parameters, eliminating the need for iterative fitting or complex inversion processes. This enables fast, accurate, and non-contact characterization of soft materials under extreme loading conditions, with significant implications for biomedical applications and materials science.
Reinforcement Learning Optimization for Large-Scale Learning: An Efficient and User-Friendly Scaling Library
Jun 06, 2025Abstract:We introduce ROLL, an efficient, scalable, and user-friendly library designed for Reinforcement Learning Optimization for Large-scale Learning. ROLL caters to three primary user groups: tech pioneers aiming for cost-effective, fault-tolerant large-scale training, developers requiring flexible control over training workflows, and researchers seeking agile experimentation. ROLL is built upon several key modules to serve these user groups effectively. First, a single-controller architecture combined with an abstraction of the parallel worker simplifies the development of the training pipeline. Second, the parallel strategy and data transfer modules enable efficient and scalable training. Third, the rollout scheduler offers fine-grained management of each sample's lifecycle during the rollout stage. Fourth, the environment worker and reward worker support rapid and flexible experimentation with agentic RL algorithms and reward designs. Finally, AutoDeviceMapping allows users to assign resources to different models flexibly across various stages.
FM-LoRA: Factorized Low-Rank Meta-Prompting for Continual Learning
Apr 09, 2025Abstract:How to adapt a pre-trained model continuously for sequential tasks with different prediction class labels and domains and finally learn a generalizable model across diverse tasks is a long-lasting challenge. Continual learning (CL) has emerged as a promising approach to leverage pre-trained models (e.g., Transformers) for sequential tasks. While many existing CL methods incrementally store additional learned structures, such as Low-Rank Adaptation (LoRA) adapters or prompts and sometimes even preserve features from previous samples to maintain performance. This leads to unsustainable parameter growth and escalating storage costs as the number of tasks increases. Moreover, current approaches often lack task similarity awareness, which further hinders the models ability to effectively adapt to new tasks without interfering with previously acquired knowledge. To address these challenges, we propose FM-LoRA, a novel and efficient low-rank adaptation method that integrates both a dynamic rank selector (DRS) and dynamic meta-prompting (DMP). This framework allocates model capacity more effectively across tasks by leveraging a shared low-rank subspace critical for preserving knowledge, thereby avoiding continual parameter expansion. Extensive experiments on various CL benchmarks, including ImageNet-R, CIFAR100, and CUB200 for class-incremental learning (CIL), and DomainNet for domain-incremental learning (DIL), with Transformers backbone demonstrate that FM-LoRA effectively mitigates catastrophic forgetting while delivering robust performance across a diverse range of tasks and domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge