Peijie Qiu
U-Harmony: Enhancing Joint Training for Segmentation Models with Universal Harmonization
Jan 21, 2026Abstract:In clinical practice, medical segmentation datasets are often limited and heterogeneous, with variations in modalities, protocols, and anatomical targets across institutions. Existing deep learning models struggle to jointly learn from such diverse data, often sacrificing either generalization or domain-specific knowledge. To overcome these challenges, we propose a joint training method called Universal Harmonization (U-Harmony), which can be integrated into deep learning-based architectures with a domain-gated head, enabling a single segmentation model to learn from heterogeneous datasets simultaneously. By integrating U-Harmony, our approach sequentially normalizes and then denormalizes feature distributions to mitigate domain-specific variations while preserving original dataset-specific knowledge. More appealingly, our framework also supports universal modality adaptation, allowing the seamless learning of new imaging modalities and anatomical classes. Extensive experiments on cross-institutional brain lesion datasets demonstrate the effectiveness of our approach, establishing a new benchmark for robust and adaptable 3D medical image segmentation models in real-world clinical settings.
AHA: Aligning Large Audio-Language Models for Reasoning Hallucinations via Counterfactual Hard Negatives
Dec 30, 2025Abstract:Although Large Audio-Language Models (LALMs) deliver state-of-the-art (SOTA) performance, they frequently suffer from hallucinations, e.g. generating text not grounded in the audio input. We analyze these grounding failures and identify a distinct taxonomy: Event Omission, False Event Identity, Temporal Relation Error, and Quantitative Temporal Error. To address this, we introduce the AHA (Audio Hallucination Alignment) framework. By leveraging counterfactual hard negative mining, our pipeline constructs a high-quality preference dataset that forces models to distinguish strict acoustic evidence from linguistically plausible fabrications. Additionally, we establish AHA-Eval, a diagnostic benchmark designed to rigorously test these fine-grained temporal reasoning capabilities. We apply this data to align Qwen2.5-Omni. The resulting model, Qwen-Audio-AHA, achieves a 13.7% improvement on AHA-Eval. Crucially, this benefit generalizes beyond our diagnostic set. Our model shows substantial gains on public benchmarks, including 1.3% on MMAU-Test and 1.6% on MMAR, outperforming latest SOTA methods.
Toward Effective Reinforcement Learning Fine-Tuning for Medical VQA in Vision-Language Models
May 20, 2025

Abstract:Recently, reinforcement learning (RL)-based tuning has shifted the trajectory of Multimodal Large Language Models (MLLMs), particularly following the introduction of Group Relative Policy Optimization (GRPO). However, directly applying it to medical tasks remains challenging for achieving clinically grounded model behavior. Motivated by the need to align model response with clinical expectations, we investigate four critical dimensions that affect the effectiveness of RL-based tuning in medical visual question answering (VQA): base model initialization strategy, the role of medical semantic alignment, the impact of length-based rewards on long-chain reasoning, and the influence of bias. We conduct extensive experiments to analyze these factors for medical MLLMs, providing new insights into how models are domain-specifically fine-tuned. Additionally, our results also demonstrate that GRPO-based RL tuning consistently outperforms standard supervised fine-tuning (SFT) in both accuracy and reasoning quality.
DRA-GRPO: Exploring Diversity-Aware Reward Adjustment for R1-Zero-Like Training of Large Language Models
May 14, 2025Abstract:Recent advances in reinforcement learning for language model post-training, such as Group Relative Policy Optimization (GRPO), have shown promise in low-resource settings. However, GRPO typically relies on solution-level and scalar reward signals that fail to capture the semantic diversity among sampled completions. This leads to what we identify as a diversity-quality inconsistency, where distinct reasoning paths may receive indistinguishable rewards. To address this limitation, we propose $\textit{Diversity-aware Reward Adjustment}$ (DRA), a method that explicitly incorporates semantic diversity into the reward computation. DRA uses Submodular Mutual Information (SMI) to downweight redundant completions and amplify rewards for diverse ones. This encourages better exploration during learning, while maintaining stable exploitation of high-quality samples. Our method integrates seamlessly with both GRPO and its variant DR.~GRPO, resulting in $\textit{DRA-GRPO}$ and $\textit{DGA-DR.~GRPO}$. We evaluate our method on five mathematical reasoning benchmarks and find that it outperforms recent strong baselines. It achieves state-of-the-art performance with an average accuracy of 58.2%, using only 7,000 fine-tuning samples and a total training cost of approximately $55. The code is available at https://github.com/xiwenc1/DRA-GRPO.
FIC-TSC: Learning Time Series Classification with Fisher Information Constraint
May 09, 2025



Abstract:Analyzing time series data is crucial to a wide spectrum of applications, including economics, online marketplaces, and human healthcare. In particular, time series classification plays an indispensable role in segmenting different phases in stock markets, predicting customer behavior, and classifying worker actions and engagement levels. These aspects contribute significantly to the advancement of automated decision-making and system optimization in real-world applications. However, there is a large consensus that time series data often suffers from domain shifts between training and test sets, which dramatically degrades the classification performance. Despite the success of (reversible) instance normalization in handling the domain shifts for time series regression tasks, its performance in classification is unsatisfactory. In this paper, we propose \textit{FIC-TSC}, a training framework for time series classification that leverages Fisher information as the constraint. We theoretically and empirically show this is an efficient and effective solution to guide the model converge toward flatter minima, which enhances its generalizability to distribution shifts. We rigorously evaluate our method on 30 UEA multivariate and 85 UCR univariate datasets. Our empirical results demonstrate the superiority of the proposed method over 14 recent state-of-the-art methods.
Talk Before You Retrieve: Agent-Led Discussions for Better RAG in Medical QA
Apr 30, 2025Abstract:Medical question answering (QA) is a reasoning-intensive task that remains challenging for large language models (LLMs) due to hallucinations and outdated domain knowledge. Retrieval-Augmented Generation (RAG) provides a promising post-training solution by leveraging external knowledge. However, existing medical RAG systems suffer from two key limitations: (1) a lack of modeling for human-like reasoning behaviors during information retrieval, and (2) reliance on suboptimal medical corpora, which often results in the retrieval of irrelevant or noisy snippets. To overcome these challenges, we propose Discuss-RAG, a plug-and-play module designed to enhance the medical QA RAG system through collaborative agent-based reasoning. Our method introduces a summarizer agent that orchestrates a team of medical experts to emulate multi-turn brainstorming, thereby improving the relevance of retrieved content. Additionally, a decision-making agent evaluates the retrieved snippets before their final integration. Experimental results on four benchmark medical QA datasets show that Discuss-RAG consistently outperforms MedRAG, especially significantly improving answer accuracy by up to 16.67% on BioASQ and 12.20% on PubMedQA. The code is available at: https://github.com/LLM-VLM-GSL/Discuss-RAG.
A BERT-Style Self-Supervised Learning CNN for Disease Identification from Retinal Images
Apr 25, 2025Abstract:In the field of medical imaging, the advent of deep learning, especially the application of convolutional neural networks (CNNs) has revolutionized the analysis and interpretation of medical images. Nevertheless, deep learning methods usually rely on large amounts of labeled data. In medical imaging research, the acquisition of high-quality labels is both expensive and difficult. The introduction of Vision Transformers (ViT) and self-supervised learning provides a pre-training strategy that utilizes abundant unlabeled data, effectively alleviating the label acquisition challenge while broadening the breadth of data utilization. However, ViT's high computational density and substantial demand for computing power, coupled with the lack of localization characteristics of its operations on image patches, limit its efficiency and applicability in many application scenarios. In this study, we employ nn-MobileNet, a lightweight CNN framework, to implement a BERT-style self-supervised learning approach. We pre-train the network on the unlabeled retinal fundus images from the UK Biobank to improve downstream application performance. We validate the results of the pre-trained model on Alzheimer's disease (AD), Parkinson's disease (PD), and various retinal diseases identification. The results show that our approach can significantly improve performance in the downstream tasks. In summary, this study combines the benefits of CNNs with the capabilities of advanced self-supervised learning in handling large-scale unlabeled data, demonstrating the potential of CNNs in the presence of label scarcity.
How Effective Can Dropout Be in Multiple Instance Learning ?
Apr 21, 2025Abstract:Multiple Instance Learning (MIL) is a popular weakly-supervised method for various applications, with a particular interest in histological whole slide image (WSI) classification. Due to the gigapixel resolution of WSI, applications of MIL in WSI typically necessitate a two-stage training scheme: first, extract features from the pre-trained backbone and then perform MIL aggregation. However, it is well-known that this suboptimal training scheme suffers from "noisy" feature embeddings from the backbone and inherent weak supervision, hindering MIL from learning rich and generalizable features. However, the most commonly used technique (i.e., dropout) for mitigating this issue has yet to be explored in MIL. In this paper, we empirically explore how effective the dropout can be in MIL. Interestingly, we observe that dropping the top-k most important instances within a bag leads to better performance and generalization even under noise attack. Based on this key observation, we propose a novel MIL-specific dropout method, termed MIL-Dropout, which systematically determines which instances to drop. Experiments on five MIL benchmark datasets and two WSI datasets demonstrate that MIL-Dropout boosts the performance of current MIL methods with a negligible computational cost. The code is available at https://github.com/ChongQingNoSubway/MILDropout.
FM-LoRA: Factorized Low-Rank Meta-Prompting for Continual Learning
Apr 09, 2025Abstract:How to adapt a pre-trained model continuously for sequential tasks with different prediction class labels and domains and finally learn a generalizable model across diverse tasks is a long-lasting challenge. Continual learning (CL) has emerged as a promising approach to leverage pre-trained models (e.g., Transformers) for sequential tasks. While many existing CL methods incrementally store additional learned structures, such as Low-Rank Adaptation (LoRA) adapters or prompts and sometimes even preserve features from previous samples to maintain performance. This leads to unsustainable parameter growth and escalating storage costs as the number of tasks increases. Moreover, current approaches often lack task similarity awareness, which further hinders the models ability to effectively adapt to new tasks without interfering with previously acquired knowledge. To address these challenges, we propose FM-LoRA, a novel and efficient low-rank adaptation method that integrates both a dynamic rank selector (DRS) and dynamic meta-prompting (DMP). This framework allocates model capacity more effectively across tasks by leveraging a shared low-rank subspace critical for preserving knowledge, thereby avoiding continual parameter expansion. Extensive experiments on various CL benchmarks, including ImageNet-R, CIFAR100, and CUB200 for class-incremental learning (CIL), and DomainNet for domain-incremental learning (DIL), with Transformers backbone demonstrate that FM-LoRA effectively mitigates catastrophic forgetting while delivering robust performance across a diverse range of tasks and domains.
Prompt-OT: An Optimal Transport Regularization Paradigm for Knowledge Preservation in Vision-Language Model Adaptation
Mar 11, 2025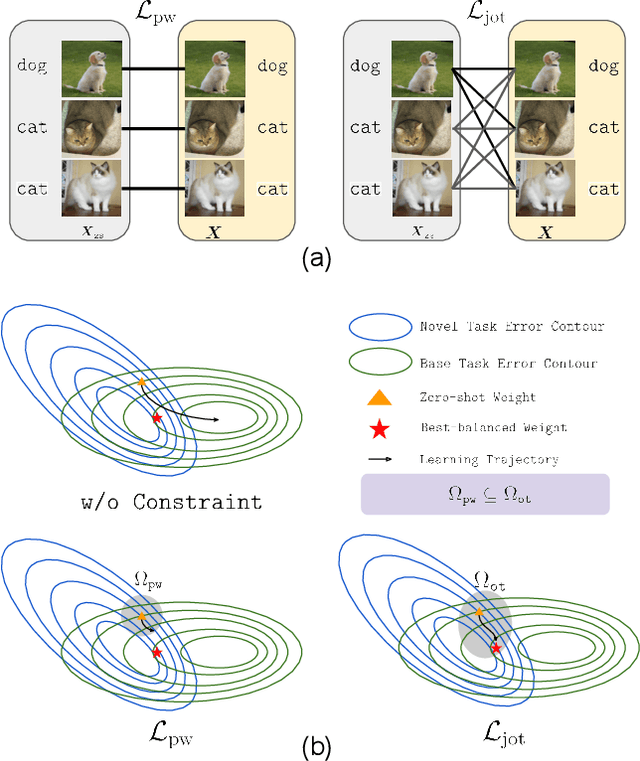



Abstract:Vision-language models (VLMs) such as CLIP demonstrate strong performance but struggle when adapted to downstream tasks. Prompt learning has emerged as an efficient and effective strategy to adapt VLMs while preserving their pre-trained knowledge. However, existing methods still lead to overfitting and degrade zero-shot generalization. To address this challenge, we propose an optimal transport (OT)-guided prompt learning framework that mitigates forgetting by preserving the structural consistency of feature distributions between pre-trained and fine-tuned models. Unlike conventional point-wise constraints, OT naturally captures cross-instance relationships and expands the feasible parameter space for prompt tuning, allowing a better trade-off between adaptation and generalization. Our approach enforces joint constraints on both vision and text representations, ensuring a holistic feature alignment. Extensive experiments on benchmark datasets demonstrate that our simple yet effective method can outperform existing prompt learning strategies in base-to-novel generalization, cross-dataset evaluation, and domain generalization without additional augmentation or ensemble techniques. The code is available at https://github.com/ChongQingNoSubway/Prompt-OT
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge