Moco V2
Papers and Code
Enhancing Contrastive Learning Inspired by the Philosophy of "The Blind Men and the Elephant"
Dec 21, 2024



Contrastive learning is a prevalent technique in self-supervised vision representation learning, typically generating positive pairs by applying two data augmentations to the same image. Designing effective data augmentation strategies is crucial for the success of contrastive learning. Inspired by the story of the blind men and the elephant, we introduce JointCrop and JointBlur. These methods generate more challenging positive pairs by leveraging the joint distribution of the two augmentation parameters, thereby enabling contrastive learning to acquire more effective feature representations. To the best of our knowledge, this is the first effort to explicitly incorporate the joint distribution of two data augmentation parameters into contrastive learning. As a plug-and-play framework without additional computational overhead, JointCrop and JointBlur enhance the performance of SimCLR, BYOL, MoCo v1, MoCo v2, MoCo v3, SimSiam, and Dino baselines with notable improvements.
Label-free Monitoring of Self-Supervised Learning Progress
Sep 10, 2024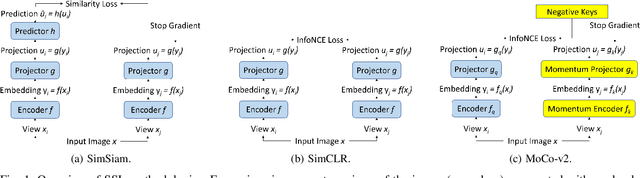
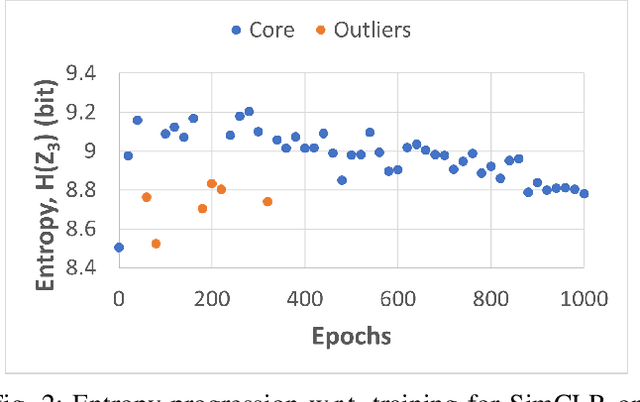
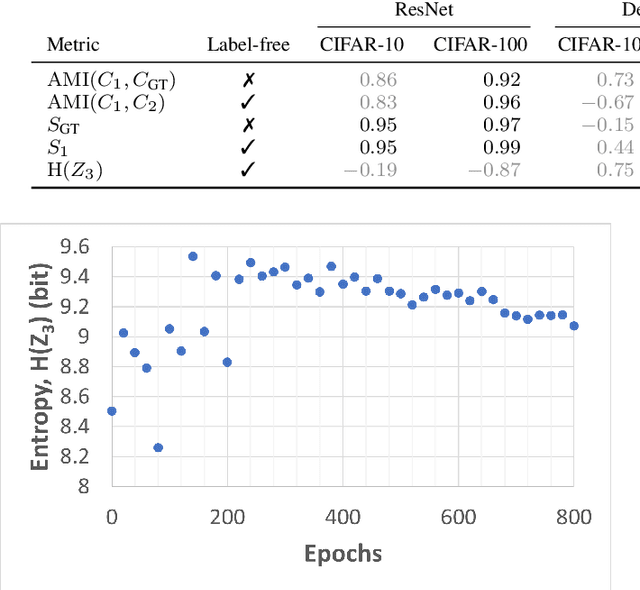
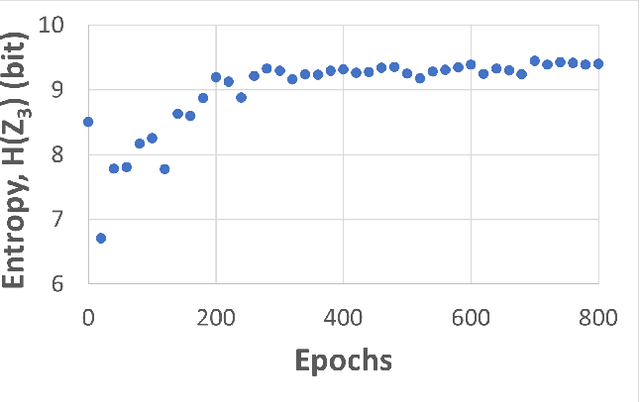
Self-supervised learning (SSL) is an effective method for exploiting unlabelled data to learn a high-level embedding space that can be used for various downstream tasks. However, existing methods to monitor the quality of the encoder -- either during training for one model or to compare several trained models -- still rely on access to annotated data. When SSL methodologies are applied to new data domains, a sufficiently large labelled dataset may not always be available. In this study, we propose several evaluation metrics which can be applied on the embeddings of unlabelled data and investigate their viability by comparing them to linear probe accuracy (a common metric which utilizes an annotated dataset). In particular, we apply $k$-means clustering and measure the clustering quality with the silhouette score and clustering agreement. We also measure the entropy of the embedding distribution. We find that while the clusters did correspond better to the ground truth annotations as training of the network progressed, label-free clustering metrics correlated with the linear probe accuracy only when training with SSL methods SimCLR and MoCo-v2, but not with SimSiam. Additionally, although entropy did not always have strong correlations with LP accuracy, this appears to be due to instability arising from early training, with the metric stabilizing and becoming more reliable at later stages of learning. Furthermore, while entropy generally decreases as learning progresses, this trend reverses for SimSiam. More research is required to establish the cause for this unexpected behaviour. Lastly, we find that while clustering based approaches are likely only viable for same-architecture comparisons, entropy may be architecture-independent.
HER2 and FISH Status Prediction in Breast Biopsy H&E-Stained Images Using Deep Learning
Aug 25, 2024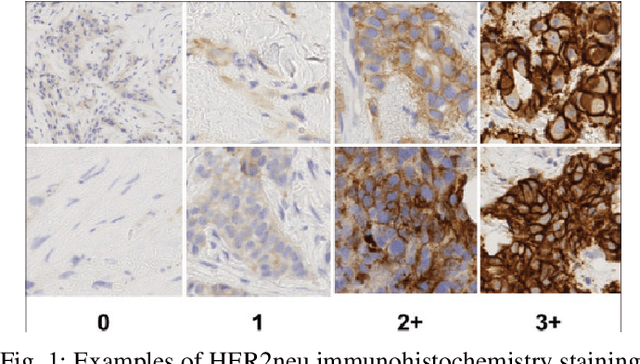
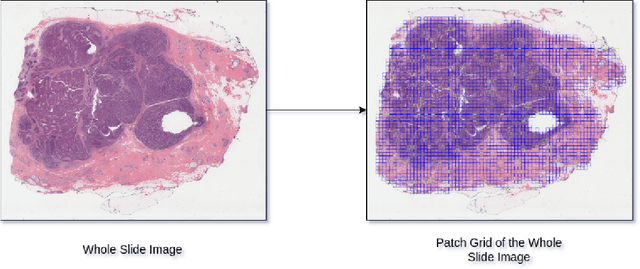

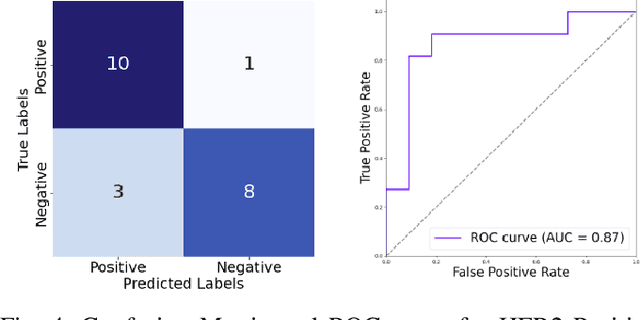
The current standard for detecting human epidermal growth factor receptor 2 (HER2) status in breast cancer patients relies on HER2 amplification, identified through fluorescence in situ hybridization (FISH) or immunohistochemistry (IHC). However, hematoxylin and eosin (H\&E) tumor stains are more widely available, and accurately predicting HER2 status using H\&E could reduce costs and expedite treatment selection. Deep Learning algorithms for H&E have shown effectiveness in predicting various cancer features and clinical outcomes, including moderate success in HER2 status prediction. In this work, we employed a customized weak supervision classification technique combined with MoCo-v2 contrastive learning to predict HER2 status. We trained our pipeline on 182 publicly available H&E Whole Slide Images (WSIs) from The Cancer Genome Atlas (TCGA), for which annotations by the pathology team at Yale School of Medicine are publicly available. Our pipeline achieved an Area Under the Curve (AUC) of 0.85 across four different test folds. Additionally, we tested our model on 44 H&E slides from the TCGA-BRCA dataset, which had an HER2 score of 2+ and included corresponding HER2 status and FISH test results. These cases are considered equivocal for IHC, requiring an expensive FISH test on their IHC slides for disambiguation. Our pipeline demonstrated an AUC of 0.81 on these challenging H&E slides. Reducing the need for FISH test can have significant implications in cancer treatment equity for underserved populations.
Contrastive Learning for Image Complexity Representation
Aug 06, 2024



Quantifying and evaluating image complexity can be instrumental in enhancing the performance of various computer vision tasks. Supervised learning can effectively learn image complexity features from well-annotated datasets. However, creating such datasets requires expensive manual annotation costs. The models may learn human subjective biases from it. In this work, we introduce the MoCo v2 framework. We utilize contrastive learning to represent image complexity, named CLIC (Contrastive Learning for Image Complexity). We find that there are complexity differences between different local regions of an image, and propose Random Crop and Mix (RCM), which can produce positive samples consisting of multi-scale local crops. RCM can also expand the train set and increase data diversity without introducing additional data. We conduct extensive experiments with CLIC, comparing it with both unsupervised and supervised methods. The results demonstrate that the performance of CLIC is comparable to that of state-of-the-art supervised methods. In addition, we establish the pipelines that can apply CLIC to computer vision tasks to effectively improve their performance.
Self-Supervised Learning Featuring Small-Scale Image Dataset for Treatable Retinal Diseases Classification
Apr 15, 2024



Automated medical diagnosis through image-based neural networks has increased in popularity and matured over years. Nevertheless, it is confined by the scarcity of medical images and the expensive labor annotation costs. Self-Supervised Learning (SSL) is an good alternative to Transfer Learning (TL) and is suitable for imbalanced image datasets. In this study, we assess four pretrained SSL models and two TL models in treatable retinal diseases classification using small-scale Optical Coherence Tomography (OCT) images ranging from 125 to 4000 with balanced or imbalanced distribution for training. The proposed SSL model achieves the state-of-art accuracy of 98.84% using only 4,000 training images. Our results suggest the SSL models provide superior performance under both the balanced and imbalanced training scenarios. The SSL model with MoCo-v2 scheme has consistent good performance under the imbalanced scenario and, especially, surpasses the other models when the training set is less than 500 images.
LeOCLR: Leveraging Original Images for Contrastive Learning of Visual Representations
Mar 11, 2024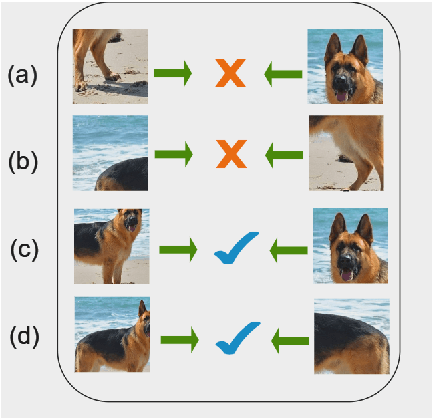
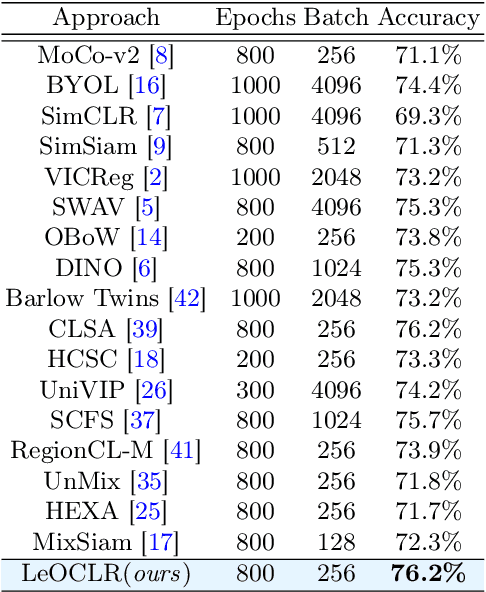
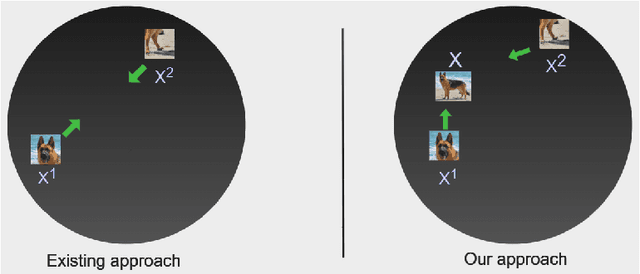
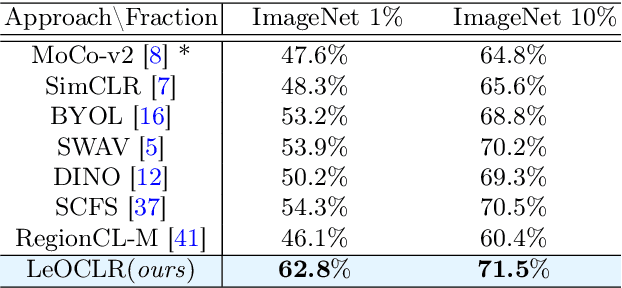
Contrastive instance discrimination outperforms supervised learning in downstream tasks like image classification and object detection. However, this approach heavily relies on data augmentation during representation learning, which may result in inferior results if not properly implemented. Random cropping followed by resizing is a common form of data augmentation used in contrastive learning, but it can lead to degraded representation learning if the two random crops contain distinct semantic content. To address this issue, this paper introduces LeOCLR (Leveraging Original Images for Contrastive Learning of Visual Representations), a framework that employs a new instance discrimination approach and an adapted loss function that ensures the shared region between positive pairs is semantically correct. The experimental results show that our approach consistently improves representation learning across different datasets compared to baseline models. For example, our approach outperforms MoCo-v2 by 5.1% on ImageNet-1K in linear evaluation and several other methods on transfer learning tasks.
Overcoming Dimensional Collapse in Self-supervised Contrastive Learning for Medical Image Segmentation
Feb 27, 2024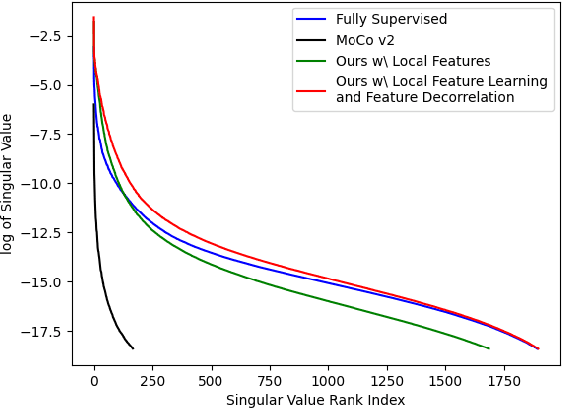
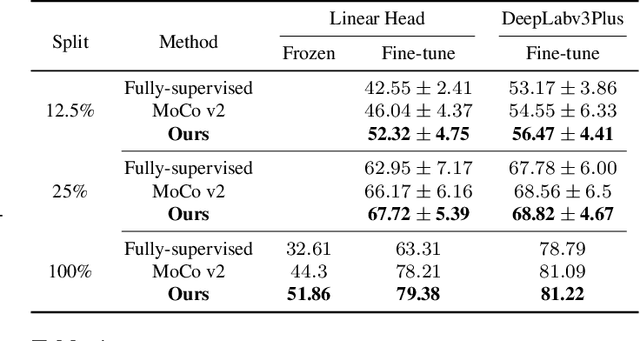
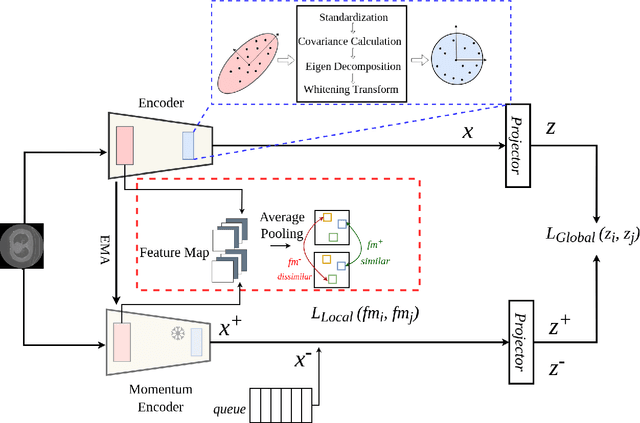
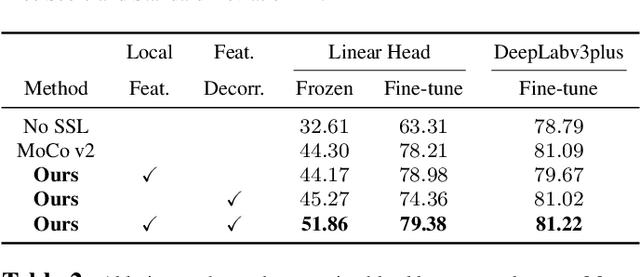
Self-supervised learning (SSL) approaches have achieved great success when the amount of labeled data is limited. Within SSL, models learn robust feature representations by solving pretext tasks. One such pretext task is contrastive learning, which involves forming pairs of similar and dissimilar input samples, guiding the model to distinguish between them. In this work, we investigate the application of contrastive learning to the domain of medical image analysis. Our findings reveal that MoCo v2, a state-of-the-art contrastive learning method, encounters dimensional collapse when applied to medical images. This is attributed to the high degree of inter-image similarity shared between the medical images. To address this, we propose two key contributions: local feature learning and feature decorrelation. Local feature learning improves the ability of the model to focus on the local regions of the image, while feature decorrelation removes the linear dependence among the features. Our experimental findings demonstrate that our contributions significantly enhance the model's performance in the downstream task of medical segmentation, both in the linear evaluation and full fine-tuning settings. This work illustrates the importance of effectively adapting SSL techniques to the characteristics of medical imaging tasks. The source code will be made publicly available at: https://github.com/CAMMA-public/med-moco
SASSL: Enhancing Self-Supervised Learning via Neural Style Transfer
Dec 02, 2023



Self-supervised learning relies heavily on data augmentation to extract meaningful representations from unlabeled images. While existing state-of-the-art augmentation pipelines incorporate a wide range of primitive transformations, these often disregard natural image structure. Thus, augmented samples can exhibit degraded semantic information and low stylistic diversity, affecting downstream performance of self-supervised representations. To overcome this, we propose SASSL: Style Augmentations for Self Supervised Learning, a novel augmentation technique based on Neural Style Transfer. The method decouples semantic and stylistic attributes in images and applies transformations exclusively to the style while preserving content, generating diverse augmented samples that better retain their semantic properties. Experimental results show our technique achieves a top-1 classification performance improvement of more than 2% on ImageNet compared to the well-established MoCo v2. We also measure transfer learning performance across five diverse datasets, observing significant improvements of up to 3.75%. Our experiments indicate that decoupling style from content information and transferring style across datasets to diversify augmentations can significantly improve downstream performance of self-supervised representations.
SegLoc: Visual Self-supervised Learning Scheme for Dense Prediction Tasks of Security Inspection X-ray Images
Oct 21, 2023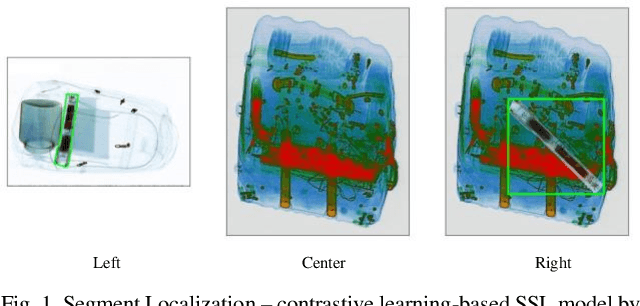

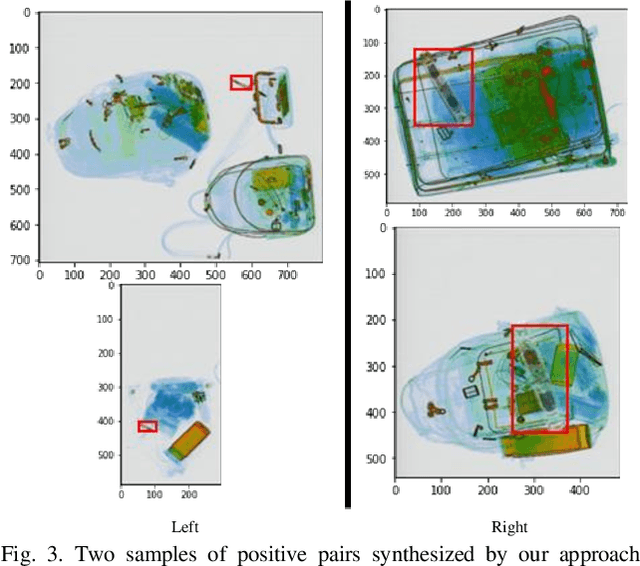
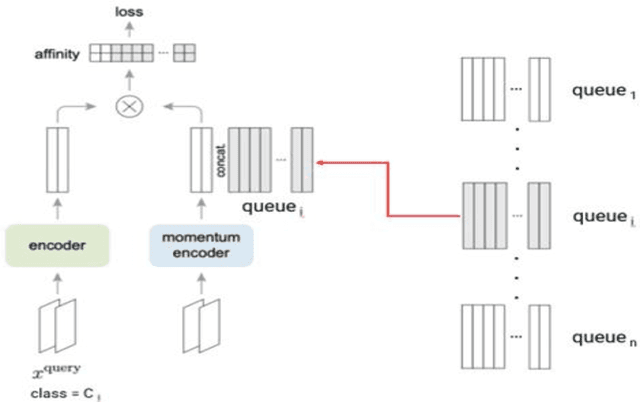
Lately, remarkable advancements of artificial intelligence have been attributed to the integration of self-supervised learning (SSL) scheme. Despite impressive achievements within natural language processing (NLP), SSL in computer vision has not been able to stay on track comparatively. Recently, integration of contrastive learning on top of existing visual SSL models has established considerable progress, thereby being able to outperform supervised counterparts. Nevertheless, the improvements were mostly limited to classification tasks; moreover, few studies have evaluated visual SSL models in real-world scenarios, while the majority considered datasets containing class-wise portrait images, notably ImageNet. Thus, here, we have considered dense prediction tasks on security inspection x-ray images to evaluate our proposed model Segmentation Localization (SegLoc). Based upon the model Instance Localization (InsLoc), our model has managed to address one of the most challenging downsides of contrastive learning, i.e., false negative pairs of query embeddings. To do so, our pre-training dataset is synthesized by cutting, transforming, then pasting labeled segments, as foregrounds, from an already existing labeled dataset (PIDray) onto instances, as backgrounds, of an unlabeled dataset (SIXray;) further, we fully harness the labels through integration of the notion, one queue per class, into MoCo-v2 memory bank, avoiding false negative pairs. Regarding the task in question, our approach has outperformed random initialization method by 3% to 6%, while having underperformed supervised initialization, in AR and AP metrics at different IoU values for 20 to 30 pre-training epochs.
Federated Model Aggregation via Self-Supervised Priors for Highly Imbalanced Medical Image Classification
Jul 27, 2023In the medical field, federated learning commonly deals with highly imbalanced datasets, including skin lesions and gastrointestinal images. Existing federated methods under highly imbalanced datasets primarily focus on optimizing a global model without incorporating the intra-class variations that can arise in medical imaging due to different populations, findings, and scanners. In this paper, we study the inter-client intra-class variations with publicly available self-supervised auxiliary networks. Specifically, we find that employing a shared auxiliary pre-trained model, like MoCo-V2, locally on every client yields consistent divergence measurements. Based on these findings, we derive a dynamic balanced model aggregation via self-supervised priors (MAS) to guide the global model optimization. Fed-MAS can be utilized with different local learning methods for effective model aggregation toward a highly robust and unbiased global model. Our code is available at \url{https://github.com/xmed-lab/Fed-MAS}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge