Yong Huang
Learning Depth from Past Selves: Self-Evolution Contrast for Robust Depth Estimation
Nov 19, 2025Abstract:Self-supervised depth estimation has gained significant attention in autonomous driving and robotics. However, existing methods exhibit substantial performance degradation under adverse weather conditions such as rain and fog, where reduced visibility critically impairs depth prediction. To address this issue, we propose a novel self-evolution contrastive learning framework called SEC-Depth for self-supervised robust depth estimation tasks. Our approach leverages intermediate parameters generated during training to construct temporally evolving latency models. Using these, we design a self-evolution contrastive scheme to mitigate performance loss under challenging conditions. Concretely, we first design a dynamic update strategy of latency models for the depth estimation task to capture optimization states across training stages. To effectively leverage latency models, we introduce a self-evolution contrastive Loss (SECL) that treats outputs from historical latency models as negative samples. This mechanism adaptively adjusts learning objectives while implicitly sensing weather degradation severity, reducing the needs for manual intervention. Experiments show that our method integrates seamlessly into diverse baseline models and significantly enhances robustness in zero-shot evaluations.
NP-LoRA: Null Space Projection Unifies Subject and Style in LoRA Fusion
Nov 14, 2025Abstract:Low-Rank Adaptation (LoRA) fusion has emerged as a key technique for reusing and composing learned subject and style representations for controllable generation without costly retraining. However, existing methods rely on weight-based merging, where one LoRA often dominates the other, leading to interference and degraded fidelity. This interference is structural: separately trained LoRAs occupy low-rank high-dimensional subspaces, leading to non-orthogonal and overlapping representations. In this work, we analyze the internal structure of LoRAs and find their generative behavior is dominated by a few principal directions in the low-rank subspace, which should remain free from interference during fusion. To achieve this, we propose Null Space Projection LoRA (NP-LoRA), a projection-based framework for LoRA fusion that enforces subspace separation to prevent structural interference among principal directions. Specifically, we first extract principal style directions via singular value decomposition (SVD) and then project the subject LoRA into its orthogonal null space. Furthermore, we introduce a soft projection mechanism that enables smooth control over the trade-off between subject fidelity and style consistency. Experiments show NP-LoRA consistently improves fusion quality over strong baselines (e.g., DINO and CLIP-based metrics, with human and LLM preference scores), and applies broadly across backbones and LoRA pairs without retraining.
Enhancing WiFi CSI Fingerprinting: A Deep Auxiliary Learning Approach
Oct 26, 2025Abstract:Radio frequency (RF) fingerprinting techniques provide a promising supplement to cryptography-based approaches but rely on dedicated equipment to capture in-phase and quadrature (IQ) samples, hindering their wide adoption. Recent advances advocate easily obtainable channel state information (CSI) by commercial WiFi devices for lightweight RF fingerprinting, while falling short in addressing the challenges of coarse granularity of CSI measurements in an open-world setting. In this paper, we propose CSI2Q, a novel CSI fingerprinting system that achieves comparable performance to IQ-based approaches. Instead of extracting fingerprints directly from raw CSI measurements, CSI2Q first transforms frequency-domain CSI measurements into time-domain signals that share the same feature space with IQ samples. Then, we employ a deep auxiliary learning strategy to transfer useful knowledge from an IQ fingerprinting model to the CSI counterpart. Finally, the trained CSI model is combined with an OpenMax function to estimate the likelihood of unknown ones. We evaluate CSI2Q on one synthetic CSI dataset involving 85 devices and two real CSI datasets, including 10 and 25 WiFi routers, respectively. Our system achieves accuracy increases of at least 16% on the synthetic CSI dataset, 20% on the in-lab CSI dataset, and 17% on the in-the-wild CSI dataset.
A Multi-Agent System for Information Extraction from the Chemical Literature
Jul 27, 2025Abstract:To fully expedite AI-powered chemical research, high-quality chemical databases are the cornerstone. Automatic extraction of chemical information from the literature is essential for constructing reaction databases, but it is currently limited by the multimodality and style variability of chemical information. In this work, we developed a multimodal large language model (MLLM)-based multi-agent system for automatic chemical information extraction. We used the MLLM's strong reasoning capability to understand the structure of complex chemical graphics, decompose the extraction task into sub-tasks and coordinate a set of specialized agents to solve them. Our system achieved an F1 score of 80.8% on a benchmark dataset of complex chemical reaction graphics from the literature, surpassing the previous state-of-the-art model (F1 score: 35.6%) by a significant margin. Additionally, it demonstrated consistent improvements in key sub-tasks, including molecular image recognition, reaction image parsing, named entity recognition and text-based reaction extraction. This work is a critical step toward automated chemical information extraction into structured datasets, which will be a strong promoter of AI-driven chemical research.
NTIRE 2025 Challenge on HR Depth from Images of Specular and Transparent Surfaces
Jun 06, 2025Abstract:This paper reports on the NTIRE 2025 challenge on HR Depth From images of Specular and Transparent surfaces, held in conjunction with the New Trends in Image Restoration and Enhancement (NTIRE) workshop at CVPR 2025. This challenge aims to advance the research on depth estimation, specifically to address two of the main open issues in the field: high-resolution and non-Lambertian surfaces. The challenge proposes two tracks on stereo and single-image depth estimation, attracting about 177 registered participants. In the final testing stage, 4 and 4 participating teams submitted their models and fact sheets for the two tracks.
SDIGLM: Leveraging Large Language Models and Multi-Modal Chain of Thought for Structural Damage Identification
Apr 12, 2025Abstract:Existing computer vision(CV)-based structural damage identification models demonstrate notable accuracy in categorizing and localizing damage. However, these models present several critical limitations that hinder their practical application in civil engineering(CE). Primarily, their ability to recognize damage types remains constrained, preventing comprehensive analysis of the highly varied and complex conditions encountered in real-world CE structures. Second, these models lack linguistic capabilities, rendering them unable to articulate structural damage characteristics through natural language descriptions. With the continuous advancement of artificial intelligence(AI), large multi-modal models(LMMs) have emerged as a transformative solution, enabling the unified encoding and alignment of textual and visual data. These models can autonomously generate detailed descriptive narratives of structural damage while demonstrating robust generalization across diverse scenarios and tasks. This study introduces SDIGLM, an innovative LMM for structural damage identification, developed based on the open-source VisualGLM-6B architecture. To address the challenge of adapting LMMs to the intricate and varied operating conditions in CE, this work integrates a U-Net-based semantic segmentation module to generate defect segmentation maps as visual Chain of Thought(CoT). Additionally, a multi-round dialogue fine-tuning dataset is constructed to enhance logical reasoning, complemented by a language CoT formed through prompt engineering. By leveraging this multi-modal CoT, SDIGLM surpasses general-purpose LMMs in structural damage identification, achieving an accuracy of 95.24% across various infrastructure types. Moreover, the model effectively describes damage characteristics such as hole size, crack direction, and corrosion severity.
Towards Large-scale Chemical Reaction Image Parsing via a Multimodal Large Language Model
Mar 11, 2025Abstract:Artificial intelligence (AI) has demonstrated significant promise in advancing organic chemistry research; however, its effectiveness depends on the availability of high-quality chemical reaction data. Currently, most published chemical reactions are not available in machine-readable form, limiting the broader application of AI in this field. The extraction of published chemical reactions into structured databases still relies heavily on manual curation, and robust automatic parsing of chemical reaction images into machine-readable data remains a significant challenge. To address this, we introduce the Reaction Image Multimodal large language model (RxnIM), the first multimodal large language model specifically designed to parse chemical reaction images into machine-readable reaction data. RxnIM not only extracts key chemical components from reaction images but also interprets the textual content that describes reaction conditions. Together with specially designed large-scale dataset generation method to support model training, our approach achieves excellent performance, with an average F1 score of 88% on various benchmarks, surpassing literature methods by 5%. This represents a crucial step toward the automatic construction of large databases of machine-readable reaction data parsed from images in the chemistry literature, providing essential data resources for AI research in chemistry. The source code, model checkpoints, and datasets developed in this work are released under permissive licenses. An instance of the RxnIM web application can be accessed at https://huggingface.co/spaces/CYF200127/RxnIM.
Molecule Generation for Target Protein Binding with Hierarchical Consistency Diffusion Model
Mar 02, 2025Abstract:Effective generation of molecular structures, or new chemical entities, that bind to target proteins is crucial for lead identification and optimization in drug discovery. Despite advancements in atom- and motif-wise deep learning models for 3D molecular generation, current methods often struggle with validity and reliability. To address these issues, we develop the Atom-Motif Consistency Diffusion Model (AMDiff), utilizing a joint-training paradigm for multi-view learning. This model features a hierarchical diffusion architecture that integrates both atom- and motif-level views of molecules, allowing for comprehensive exploration of complementary information. By leveraging classifier-free guidance and incorporating binding site features as conditional inputs, AMDiff ensures robust molecule generation across diverse targets. Compared to existing approaches, AMDiff exhibits superior validity and novelty in generating molecules tailored to fit various protein pockets. Case studies targeting protein kinases, including Anaplastic Lymphoma Kinase (ALK) and Cyclin-dependent kinase 4 (CDK4), demonstrate the model's capability in structure-based de novo drug design. Overall, AMDiff bridges the gap between atom-view and motif-view drug discovery and speeds up the process of target-aware molecular generation.
MoR: Mixture of Ranks for Low-Rank Adaptation Tuning
Oct 17, 2024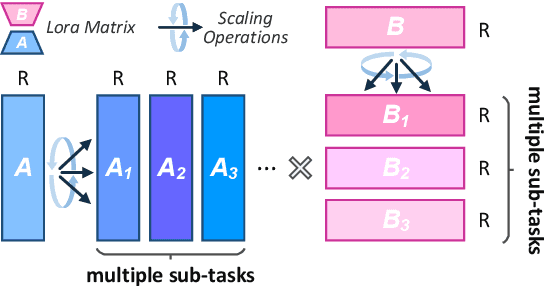
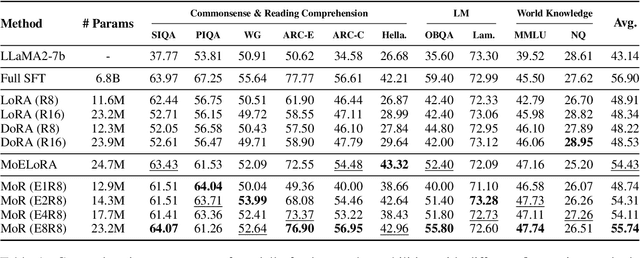
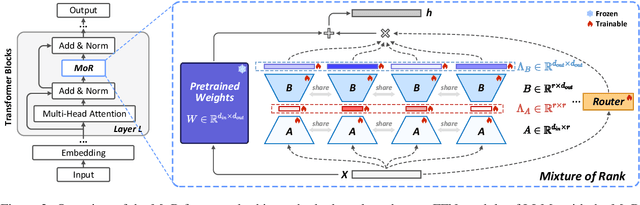
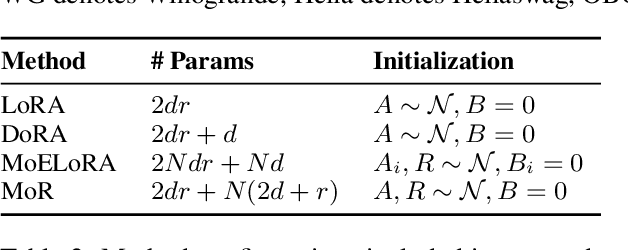
Abstract:Low-Rank Adaptation (LoRA) drives research to align its performance with full fine-tuning. However, significant challenges remain: (1) Simply increasing the rank size of LoRA does not effectively capture high-rank information, which leads to a performance bottleneck.(2) MoE-style LoRA methods substantially increase parameters and inference latency, contradicting the goals of efficient fine-tuning and ease of application. To address these challenges, we introduce Mixture of Ranks (MoR), which learns rank-specific information for different tasks based on input and efficiently integrates multi-rank information. We firstly propose a new framework that equates the integration of multiple LoRAs to expanding the rank of LoRA. Moreover, we hypothesize that low-rank LoRA already captures sufficient intrinsic information, and MoR can derive high-rank information through mathematical transformations of the low-rank components. Thus, MoR can reduces the learning difficulty of LoRA and enhances its multi-task capabilities. MoR achieves impressive results, with MoR delivering a 1.31\% performance improvement while using only 93.93\% of the parameters compared to baseline methods.
MolNexTR: A Generalized Deep Learning Model for Molecular Image Recognition
Mar 08, 2024



Abstract:In the field of chemical structure recognition, the task of converting molecular images into graph structures and SMILES string stands as a significant challenge, primarily due to the varied drawing styles and conventions prevalent in chemical literature. To bridge this gap, we proposed MolNexTR, a novel image-to-graph deep learning model that collaborates to fuse the strengths of ConvNext, a powerful Convolutional Neural Network variant, and Vision-TRansformer. This integration facilitates a more nuanced extraction of both local and global features from molecular images. MolNexTR can predict atoms and bonds simultaneously and understand their layout rules. It also excels at flexibly integrating symbolic chemistry principles to discern chirality and decipher abbreviated structures. We further incorporate a series of advanced algorithms, including improved data augmentation module, image contamination module, and a post-processing module to get the final SMILES output. These modules synergistically enhance the model's robustness against the diverse styles of molecular imagery found in real literature. In our test sets, MolNexTR has demonstrated superior performance, achieving an accuracy rate of 81-97%, marking a significant advancement in the domain of molecular structure recognition. Scientific contribution: MolNexTR is a novel image-to-graph model that incorporates a unique dual-stream encoder to extract complex molecular image features, and combines chemical rules to predict atoms and bonds while understanding atom and bond layout rules. In addition, it employs a series of novel augmentation algorithms to significantly enhance the robustness and performance of the model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge