Yindalon Aphinyanaphongs
Generalist Foundation Models Are Not Clinical Enough for Hospital Operations
Nov 17, 2025Abstract:Hospitals and healthcare systems rely on operational decisions that determine patient flow, cost, and quality of care. Despite strong performance on medical knowledge and conversational benchmarks, foundation models trained on general text may lack the specialized knowledge required for these operational decisions. We introduce Lang1, a family of models (100M-7B parameters) pretrained on a specialized corpus blending 80B clinical tokens from NYU Langone Health's EHRs and 627B tokens from the internet. To rigorously evaluate Lang1 in real-world settings, we developed the REalistic Medical Evaluation (ReMedE), a benchmark derived from 668,331 EHR notes that evaluates five critical tasks: 30-day readmission prediction, 30-day mortality prediction, length of stay, comorbidity coding, and predicting insurance claims denial. In zero-shot settings, both general-purpose and specialized models underperform on four of five tasks (36.6%-71.7% AUROC), with mortality prediction being an exception. After finetuning, Lang1-1B outperforms finetuned generalist models up to 70x larger and zero-shot models up to 671x larger, improving AUROC by 3.64%-6.75% and 1.66%-23.66% respectively. We also observed cross-task scaling with joint finetuning on multiple tasks leading to improvement on other tasks. Lang1-1B effectively transfers to out-of-distribution settings, including other clinical tasks and an external health system. Our findings suggest that predictive capabilities for hospital operations require explicit supervised finetuning, and that this finetuning process is made more efficient by in-domain pretraining on EHR. Our findings support the emerging view that specialized LLMs can compete with generalist models in specialized tasks, and show that effective healthcare systems AI requires the combination of in-domain pretraining, supervised finetuning, and real-world evaluation beyond proxy benchmarks.
Repurposing the scientific literature with vision-language models
Feb 26, 2025Abstract:Research in AI for Science often focuses on using AI technologies to augment components of the scientific process, or in some cases, the entire scientific method; how about AI for scientific publications? Peer-reviewed journals are foundational repositories of specialized knowledge, written in discipline-specific language that differs from general Internet content used to train most large language models (LLMs) and vision-language models (VLMs). We hypothesized that by combining a family of scientific journals with generative AI models, we could invent novel tools for scientific communication, education, and clinical care. We converted 23,000 articles from Neurosurgery Publications into a multimodal database - NeuroPubs - of 134 million words and 78,000 image-caption pairs to develop six datasets for building AI models. We showed that the content of NeuroPubs uniquely represents neurosurgery-specific clinical contexts compared with broader datasets and PubMed. For publishing, we employed generalist VLMs to automatically generate graphical abstracts from articles. Editorial board members rated 70% of these as ready for publication without further edits. For education, we generated 89,587 test questions in the style of the ABNS written board exam, which trainee and faculty neurosurgeons found indistinguishable from genuine examples 54% of the time. We used these questions alongside a curriculum learning process to track knowledge acquisition while training our 34 billion-parameter VLM (CNS-Obsidian). In a blinded, randomized controlled trial, we demonstrated the non-inferiority of CNS-Obsidian to GPT-4o (p = 0.1154) as a diagnostic copilot for a neurosurgical service. Our findings lay a novel foundation for AI with Science and establish a framework to elevate scientific communication using state-of-the-art generative artificial intelligence while maintaining rigorous quality standards.
Generalization in Healthcare AI: Evaluation of a Clinical Large Language Model
Feb 24, 2024Abstract:Advances in large language models (LLMs) provide new opportunities in healthcare for improved patient care, clinical decision-making, and enhancement of physician and administrator workflows. However, the potential of these models importantly depends on their ability to generalize effectively across clinical environments and populations, a challenge often underestimated in early development. To better understand reasons for these challenges and inform mitigation approaches, we evaluated ClinicLLM, an LLM trained on [HOSPITAL]'s clinical notes, analyzing its performance on 30-day all-cause readmission prediction focusing on variability across hospitals and patient characteristics. We found poorer generalization particularly in hospitals with fewer samples, among patients with government and unspecified insurance, the elderly, and those with high comorbidities. To understand reasons for lack of generalization, we investigated sample sizes for fine-tuning, note content (number of words per note), patient characteristics (comorbidity level, age, insurance type, borough), and health system aspects (hospital, all-cause 30-day readmission, and mortality rates). We used descriptive statistics and supervised classification to identify features. We found that, along with sample size, patient age, number of comorbidities, and the number of words in notes are all important factors related to generalization. Finally, we compared local fine-tuning (hospital specific), instance-based augmented fine-tuning and cluster-based fine-tuning for improving generalization. Among these, local fine-tuning proved most effective, increasing AUC by 0.25% to 11.74% (most helpful in settings with limited data). Overall, this study provides new insights for enhancing the deployment of large language models in the societally important domain of healthcare, and improving their performance for broader populations.
A dynamic risk score for early prediction of cardiogenic shock using machine learning
Mar 28, 2023



Abstract:Myocardial infarction and heart failure are major cardiovascular diseases that affect millions of people in the US. The morbidity and mortality are highest among patients who develop cardiogenic shock. Early recognition of cardiogenic shock is critical. Prompt implementation of treatment measures can prevent the deleterious spiral of ischemia, low blood pressure, and reduced cardiac output due to cardiogenic shock. However, early identification of cardiogenic shock has been challenging due to human providers' inability to process the enormous amount of data in the cardiac intensive care unit (ICU) and lack of an effective risk stratification tool. We developed a deep learning-based risk stratification tool, called CShock, for patients admitted into the cardiac ICU with acute decompensated heart failure and/or myocardial infarction to predict onset of cardiogenic shock. To develop and validate CShock, we annotated cardiac ICU datasets with physician adjudicated outcomes. CShock achieved an area under the receiver operator characteristic curve (AUROC) of 0.820, which substantially outperformed CardShock (AUROC 0.519), a well-established risk score for cardiogenic shock prognosis. CShock was externally validated in an independent patient cohort and achieved an AUROC of 0.800, demonstrating its generalizability in other cardiac ICUs.
New-Onset Diabetes Assessment Using Artificial Intelligence-Enhanced Electrocardiography
May 05, 2022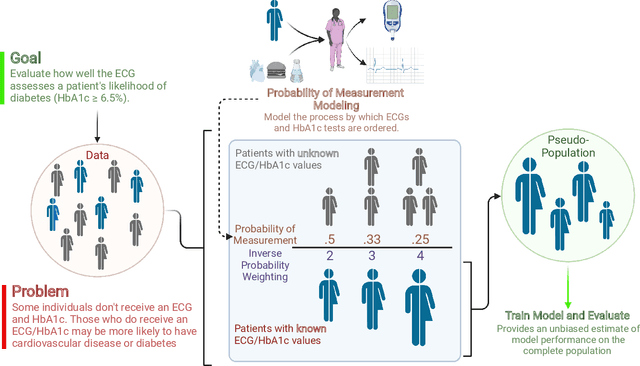
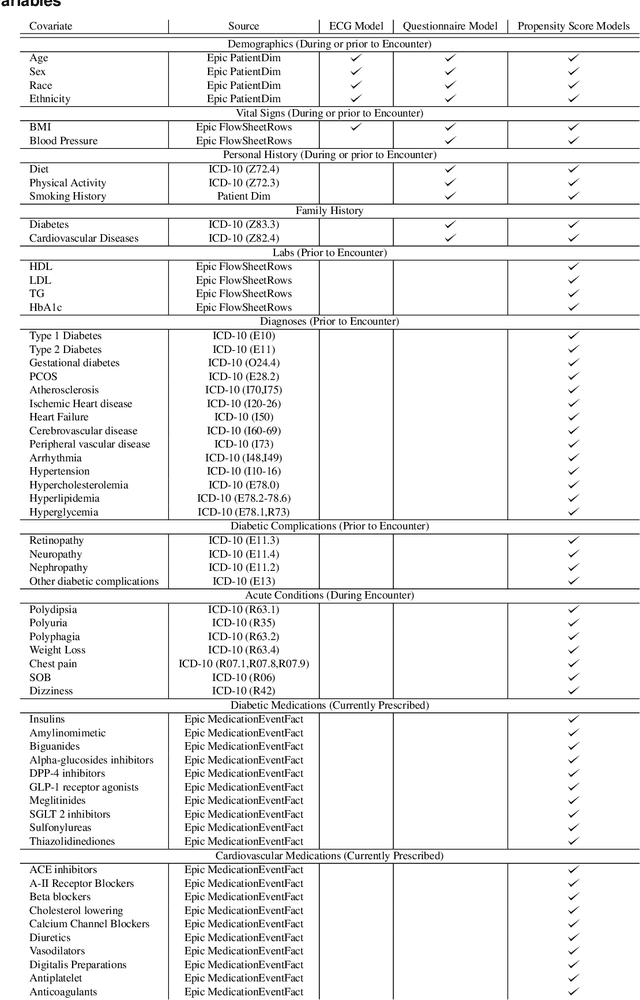
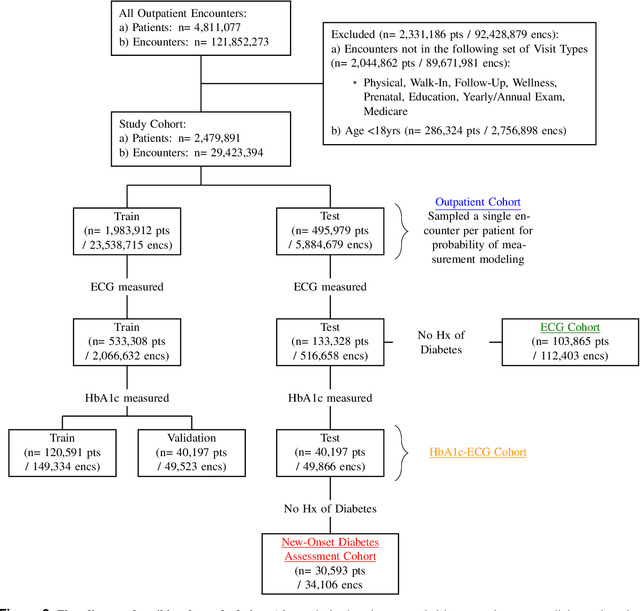
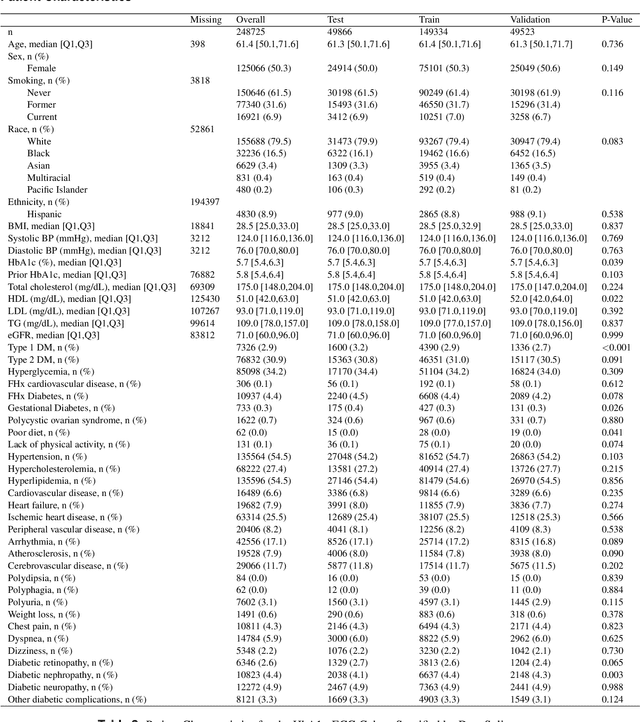
Abstract:Undiagnosed diabetes is present in 21.4% of adults with diabetes. Diabetes can remain asymptomatic and undetected due to limitations in screening rates. To address this issue, questionnaires, such as the American Diabetes Association (ADA) Risk test, have been recommended for use by physicians and the public. Based on evidence that blood glucose concentration can affect cardiac electrophysiology, we hypothesized that an artificial intelligence (AI)-enhanced electrocardiogram (ECG) could identify adults with new-onset diabetes. We trained a neural network to estimate HbA1c using a 12-lead ECG and readily available demographics. We retrospectively assembled a dataset comprised of patients with paired ECG and HbA1c data. The population of patients who receive both an ECG and HbA1c may a biased sample of the complete outpatient population, so we adjusted the importance placed on each patient to generate a more representative pseudo-population. We found ECG-based assessment outperforms the ADA Risk test, achieving a higher area under the curve (0.80 vs. 0.68) and positive predictive value (14% vs. 9%) -- 2.6 times the prevalence of diabetes in the cohort. The AI-enhanced ECG significantly outperforms electrophysiologist interpretation of the ECG, suggesting that the task is beyond current clinical capabilities. Given the prevalence of ECGs in clinics and via wearable devices, such a tool would make precise, automated diabetes assessment widely accessible.
Have We Learned to Explain?: How Interpretability Methods Can Learn to Encode Predictions in their Interpretations
Mar 02, 2021
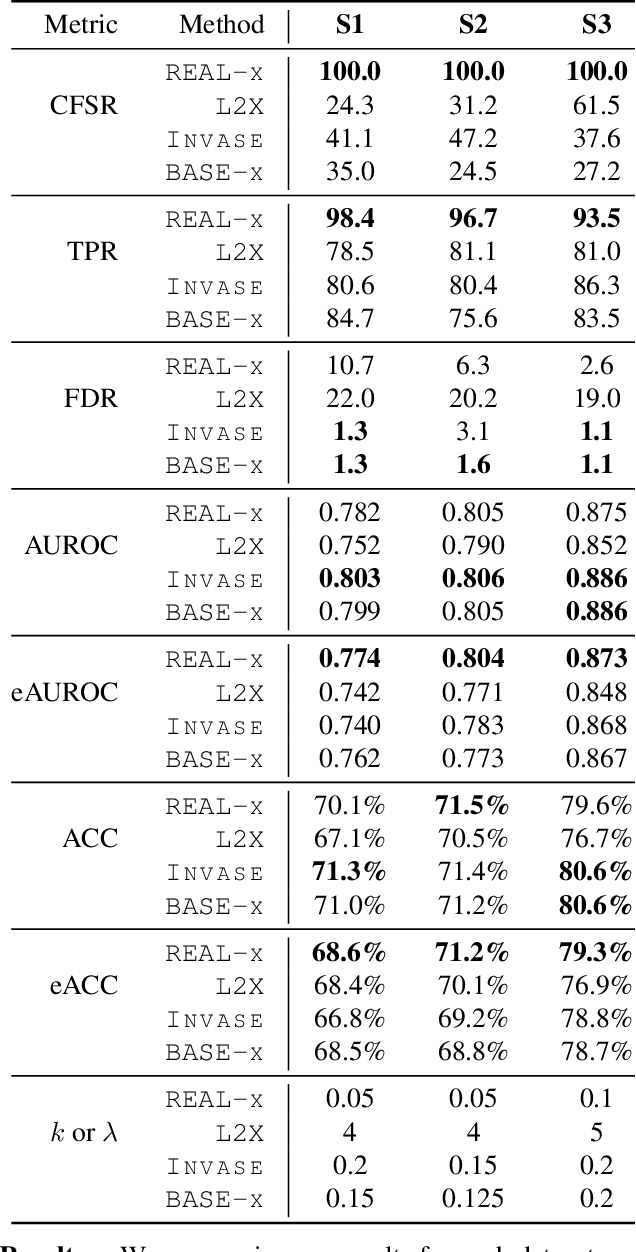
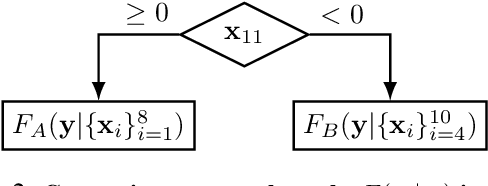
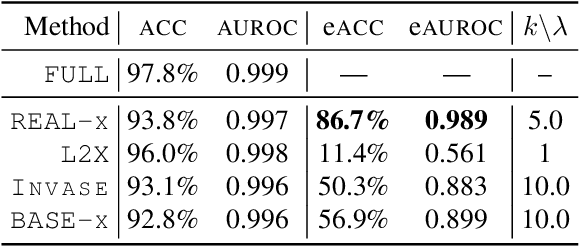
Abstract:While the need for interpretable machine learning has been established, many common approaches are slow, lack fidelity, or hard to evaluate. Amortized explanation methods reduce the cost of providing interpretations by learning a global selector model that returns feature importances for a single instance of data. The selector model is trained to optimize the fidelity of the interpretations, as evaluated by a predictor model for the target. Popular methods learn the selector and predictor model in concert, which we show allows predictions to be encoded within interpretations. We introduce EVAL-X as a method to quantitatively evaluate interpretations and REAL-X as an amortized explanation method, which learn a predictor model that approximates the true data generating distribution given any subset of the input. We show EVAL-X can detect when predictions are encoded in interpretations and show the advantages of REAL-X through quantitative and radiologist evaluation.
* 15 pages, 3 figures, Proceedings of the 24th International Conference on Artificial Intelligence and Statistics (AISTATS) 2021
COVID-19 Prognosis via Self-Supervised Representation Learning and Multi-Image Prediction
Jan 25, 2021
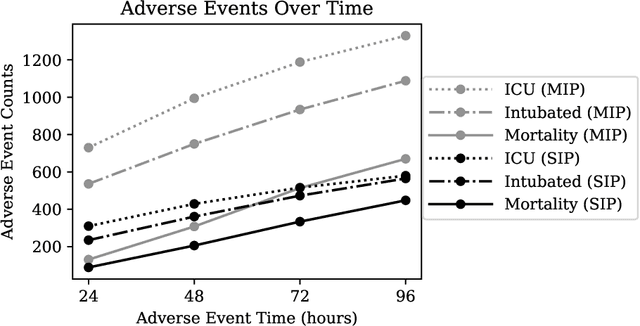
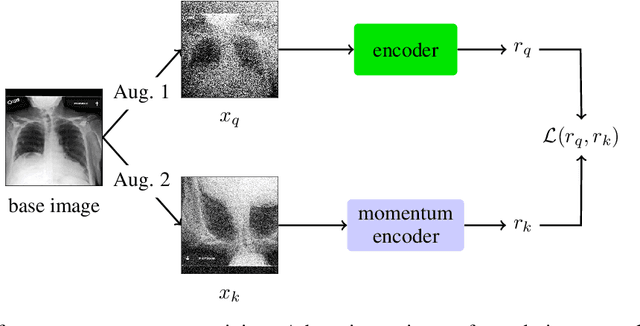
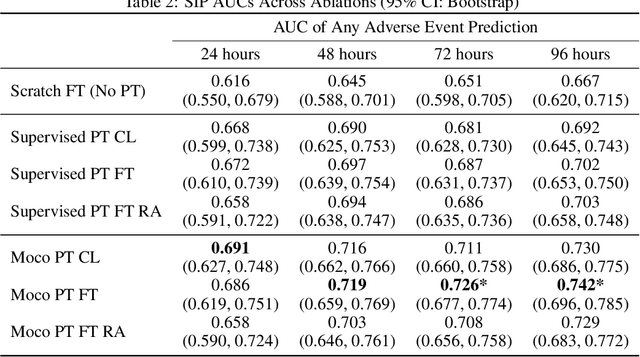
Abstract:The rapid spread of COVID-19 cases in recent months has strained hospital resources, making rapid and accurate triage of patients presenting to emergency departments a necessity. Machine learning techniques using clinical data such as chest X-rays have been used to predict which patients are most at risk of deterioration. We consider the task of predicting two types of patient deterioration based on chest X-rays: adverse event deterioration (i.e., transfer to the intensive care unit, intubation, or mortality) and increased oxygen requirements beyond 6 L per day. Due to the relative scarcity of COVID-19 patient data, existing solutions leverage supervised pretraining on related non-COVID images, but this is limited by the differences between the pretraining data and the target COVID-19 patient data. In this paper, we use self-supervised learning based on the momentum contrast (MoCo) method in the pretraining phase to learn more general image representations to use for downstream tasks. We present three results. The first is deterioration prediction from a single image, where our model achieves an area under receiver operating characteristic curve (AUC) of 0.742 for predicting an adverse event within 96 hours (compared to 0.703 with supervised pretraining) and an AUC of 0.765 for predicting oxygen requirements greater than 6 L a day at 24 hours (compared to 0.749 with supervised pretraining). We then propose a new transformer-based architecture that can process sequences of multiple images for prediction and show that this model can achieve an improved AUC of 0.786 for predicting an adverse event at 96 hours and an AUC of 0.848 for predicting mortalities at 96 hours. A small pilot clinical study suggested that the prediction accuracy of our model is comparable to that of experienced radiologists analyzing the same information.
An artificial intelligence system for predicting the deterioration of COVID-19 patients in the emergency department
Aug 04, 2020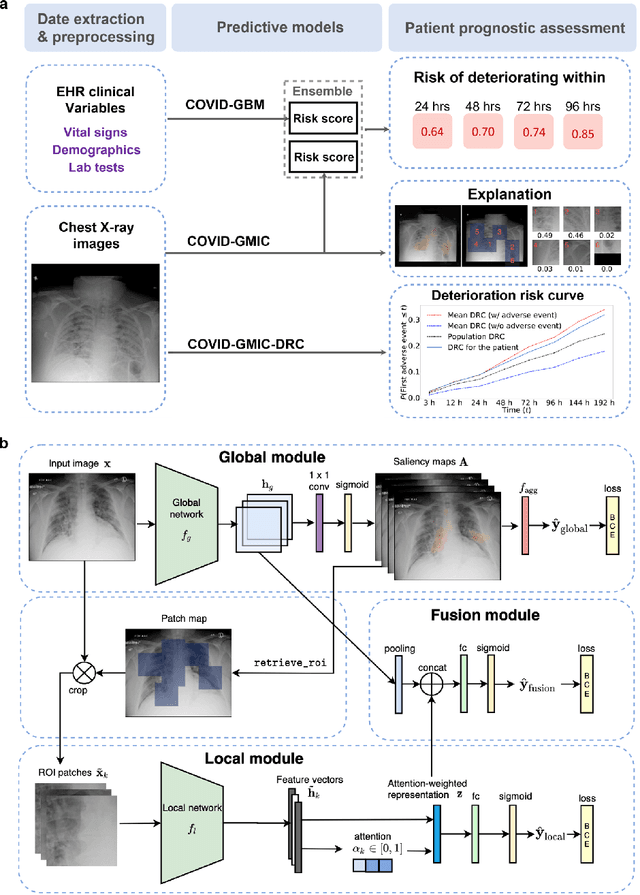
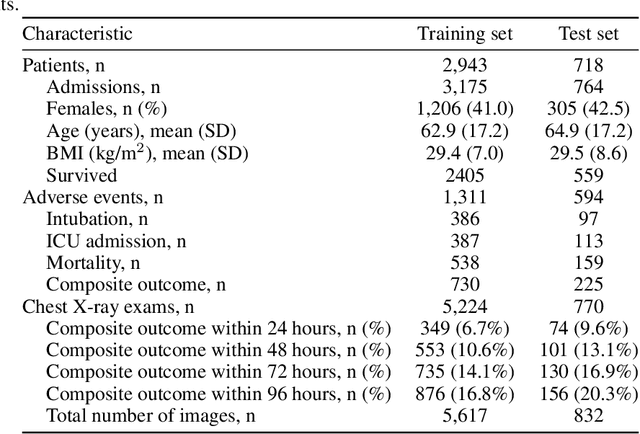
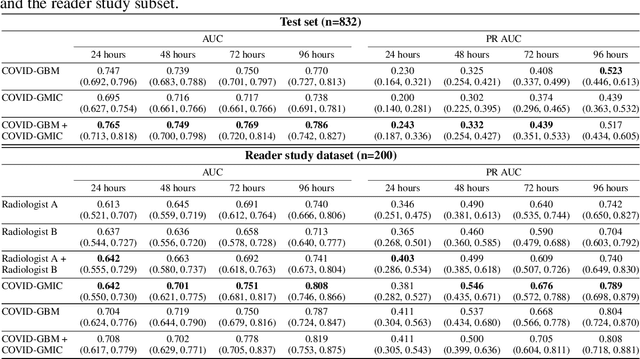
Abstract:During the COVID-19 pandemic, rapid and accurate triage of patients at the emergency department is critical to inform decision-making. We propose a data-driven approach for automatic prediction of deterioration risk using a deep neural network that learns from chest X-ray images, and a gradient boosting model that learns from routine clinical variables. Our AI prognosis system, trained using data from 3,661 patients, achieves an AUC of 0.786 (95% CI: 0.742-0.827) when predicting deterioration within 96 hours. The deep neural network extracts informative areas of chest X-ray images to assist clinicians in interpreting the predictions, and performs comparably to two radiologists in a reader study. In order to verify performance in a real clinical setting, we silently deployed a preliminary version of the deep neural network at NYU Langone Health during the first wave of the pandemic, which produced accurate predictions in real-time. In summary, our findings demonstrate the potential of the proposed system for assisting front-line physicians in the triage of COVID-19 patients.
Assessment of Amazon Comprehend Medical: Medication Information Extraction
Feb 02, 2020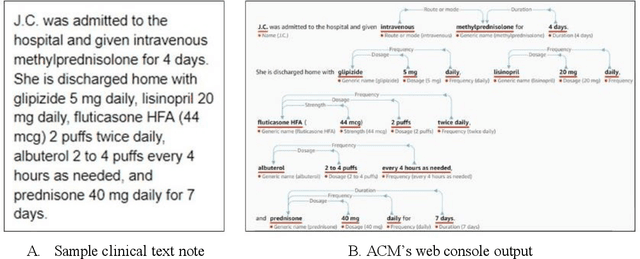
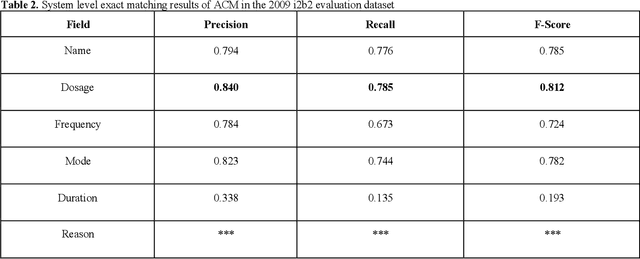
Abstract:In November 27, 2018, Amazon Web Services (AWS) released Amazon Comprehend Medical (ACM), a deep learning based system that automatically extracts clinical concepts (which include anatomy, medical conditions, protected health information (PH)I, test names, treatment names, and medical procedures, and medications) from clinical text notes. Uptake and trust in any new data product relies on independent validation across benchmark datasets and tools to establish and confirm expected quality of results. This work focuses on the medication extraction task, and particularly, ACM was evaluated using the official test sets from the 2009 i2b2 Medication Extraction Challenge and 2018 n2c2 Track 2: Adverse Drug Events and Medication Extraction in EHRs. Overall, ACM achieved F-scores of 0.768 and 0.828. These scores ranked the lowest when compared to the three best systems in the respective challenges. To further establish the generalizability of its medication extraction performance, a set of random internal clinical text notes from NYU Langone Medical Center were also included in this work. And in this corpus, ACM garnered an F-score of 0.753.
Utility of General and Specific Word Embeddings for Classifying Translational Stages of Research
Jul 09, 2018

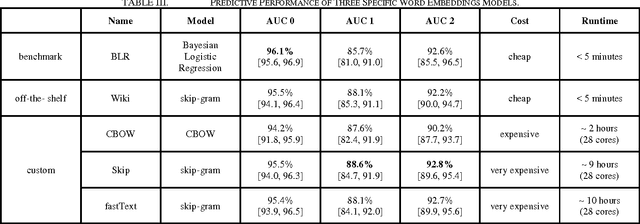
Abstract:Conventional text classification models make a bag-of-words assumption reducing text into word occurrence counts per document. Recent algorithms such as word2vec are capable of learning semantic meaning and similarity between words in an entirely unsupervised manner using a contextual window and doing so much faster than previous methods. Each word is projected into vector space such that similar meaning words such as "strong" and "powerful" are projected into the same general Euclidean space. Open questions about these embeddings include their utility across classification tasks and the optimal properties and source of documents to construct broadly functional embeddings. In this work, we demonstrate the usefulness of pre-trained embeddings for classification in our task and demonstrate that custom word embeddings, built in the domain and for the tasks, can improve performance over word embeddings learnt on more general data including news articles or Wikipedia.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge