Jordan Swartz
CLIP: A Dataset for Extracting Action Items for Physicians from Hospital Discharge Notes
Jun 04, 2021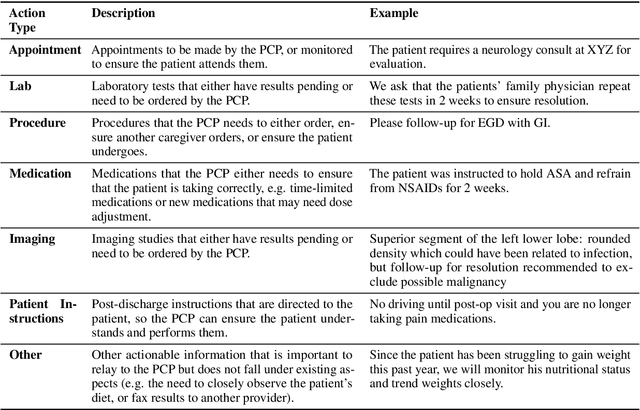
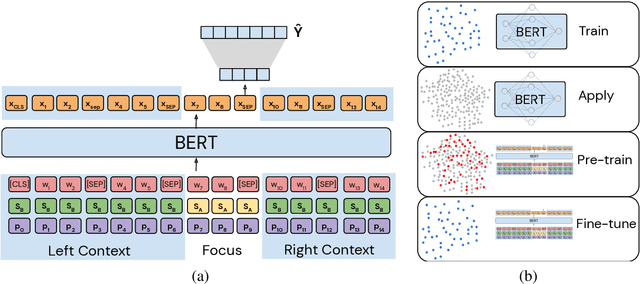
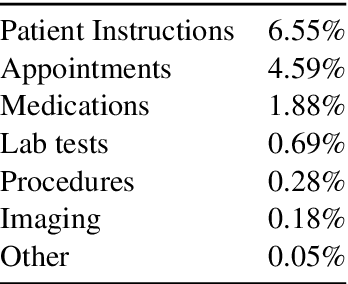
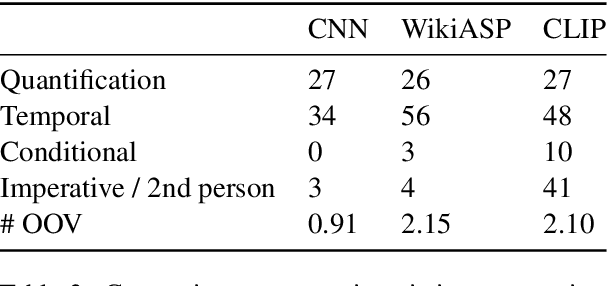
Abstract:Continuity of care is crucial to ensuring positive health outcomes for patients discharged from an inpatient hospital setting, and improved information sharing can help. To share information, caregivers write discharge notes containing action items to share with patients and their future caregivers, but these action items are easily lost due to the lengthiness of the documents. In this work, we describe our creation of a dataset of clinical action items annotated over MIMIC-III, the largest publicly available dataset of real clinical notes. This dataset, which we call CLIP, is annotated by physicians and covers 718 documents representing 100K sentences. We describe the task of extracting the action items from these documents as multi-aspect extractive summarization, with each aspect representing a type of action to be taken. We evaluate several machine learning models on this task, and show that the best models exploit in-domain language model pre-training on 59K unannotated documents, and incorporate context from neighboring sentences. We also propose an approach to pre-training data selection that allows us to explore the trade-off between size and domain-specificity of pre-training datasets for this task.
Knowledge Base Completion for Constructing Problem-Oriented Medical Records
Apr 27, 2020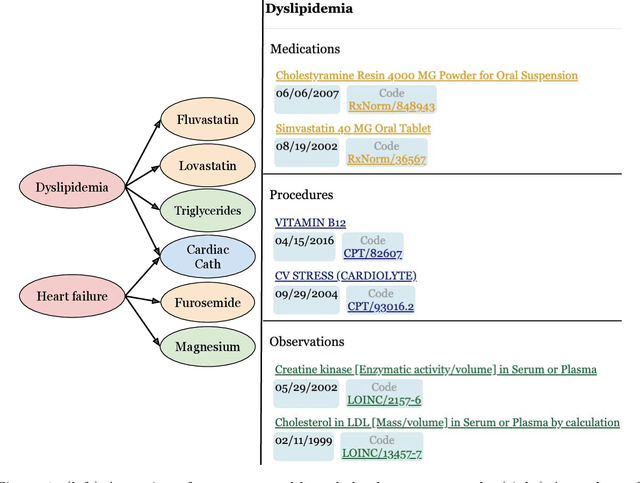
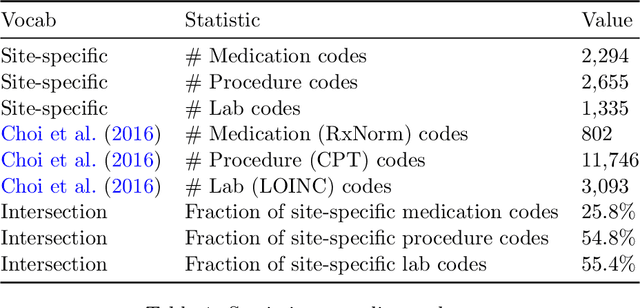
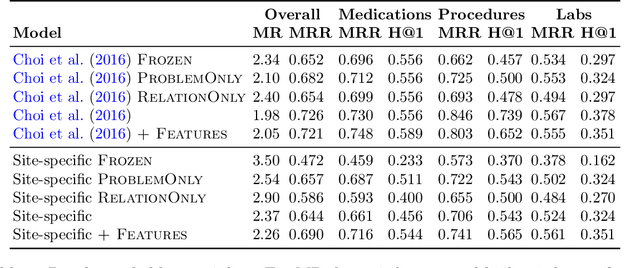
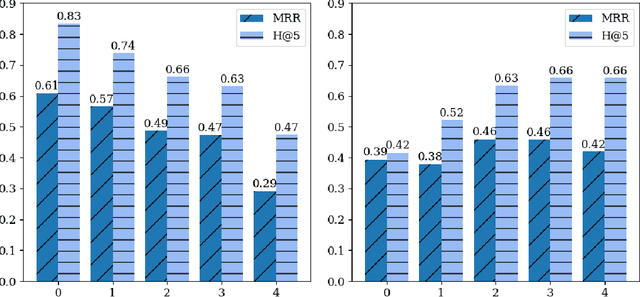
Abstract:Both electronic health records and personal health records are typically organized by data type, with medical problems, medications, procedures, and laboratory results chronologically sorted in separate areas of the chart. As a result, it can be difficult to find all of the relevant information for answering a clinical question about a given medical problem. A promising alternative is to instead organize by problems, with related medications, procedures, and other pertinent information all grouped together. A recent effort by Buchanan (2017) manually defined, through expert consensus, 11 medical problems and the relevant labs and medications for each. We show how to use machine learning on electronic health records to instead automatically construct these problem-based groupings of relevant medications, procedures, and laboratory tests. We formulate the learning task as one of knowledge base completion, and annotate a dataset that expands the set of problems from 11 to 32. We develop a model architecture that exploits both pre-trained concept embeddings and usage data relating the concepts contained in a longitudinal dataset from a large health system. We evaluate our algorithms' ability to suggest relevant medications, procedures, and lab tests, and find that the approach provides feasible suggestions even for problems that are hidden during training.
A Workflow for Visual Diagnostics of Binary Classifiers using Instance-Level Explanations
Oct 01, 2017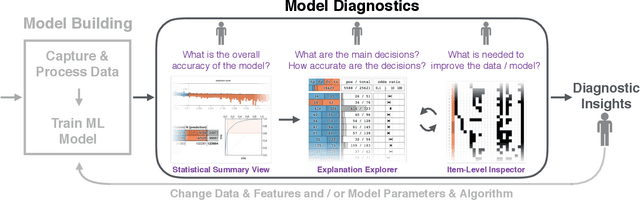
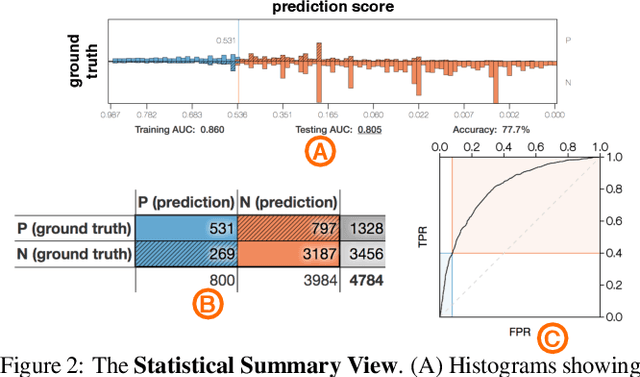
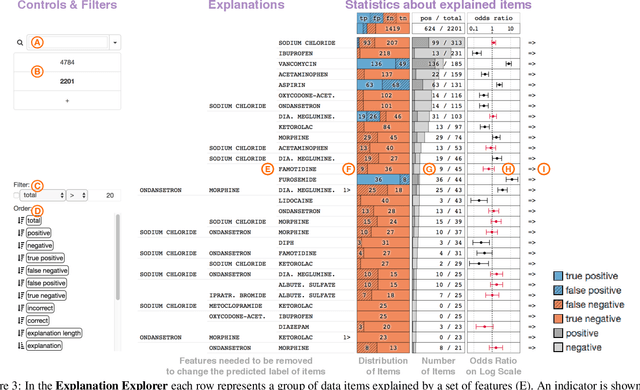
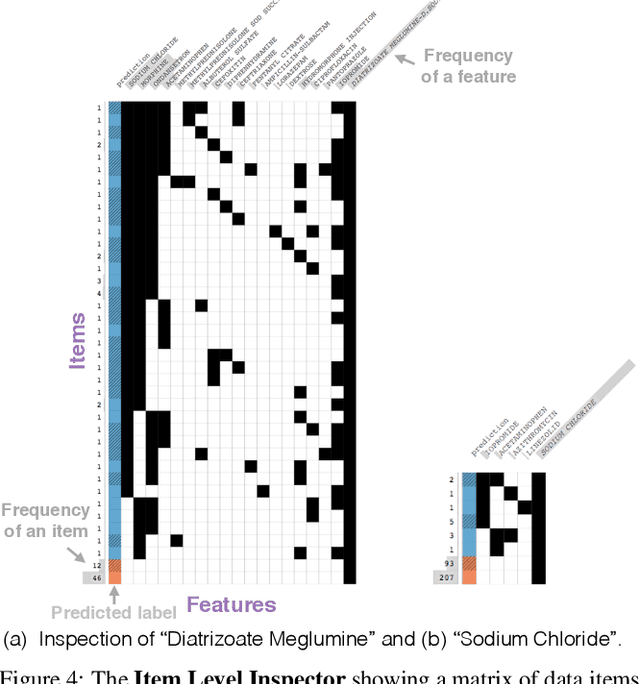
Abstract:Human-in-the-loop data analysis applications necessitate greater transparency in machine learning models for experts to understand and trust their decisions. To this end, we propose a visual analytics workflow to help data scientists and domain experts explore, diagnose, and understand the decisions made by a binary classifier. The approach leverages "instance-level explanations", measures of local feature relevance that explain single instances, and uses them to build a set of visual representations that guide the users in their investigation. The workflow is based on three main visual representations and steps: one based on aggregate statistics to see how data distributes across correct / incorrect decisions; one based on explanations to understand which features are used to make these decisions; and one based on raw data, to derive insights on potential root causes for the observed patterns. The workflow is derived from a long-term collaboration with a group of machine learning and healthcare professionals who used our method to make sense of machine learning models they developed. The case study from this collaboration demonstrates that the proposed workflow helps experts derive useful knowledge about the model and the phenomena it describes, thus experts can generate useful hypotheses on how a model can be improved.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge