Weibin Liao
HyFunc: Accelerating LLM-based Function Calls for Agentic AI through Hybrid-Model Cascade and Dynamic Templating
Feb 14, 2026Abstract:While agentic AI systems rely on LLMs to translate user intent into structured function calls, this process is fraught with computational redundancy, leading to high inference latency that hinders real-time applications. This paper identifies and addresses three key redundancies: (1) the redundant processing of a large library of function descriptions for every request; (2) the redundant use of a large, slow model to generate an entire, often predictable, token sequence; and (3) the redundant generation of fixed, boilerplate parameter syntax. We introduce HyFunc, a novel framework that systematically eliminates these inefficiencies. HyFunc employs a hybrid-model cascade where a large model distills user intent into a single "soft token." This token guides a lightweight retriever to select relevant functions and directs a smaller, prefix-tuned model to generate the final call, thus avoiding redundant context processing and full-sequence generation by the large model. To eliminate syntactic redundancy, our "dynamic templating" technique injects boilerplate parameter syntax on-the-fly within an extended vLLM engine. To avoid potential limitations in generalization, we evaluate HyFunc on an unseen benchmark dataset, BFCL. Experimental results demonstrate that HyFunc achieves an excellent balance between efficiency and performance. It achieves an inference latency of 0.828 seconds, outperforming all baseline models, and reaches a performance of 80.1%, surpassing all models with a comparable parameter scale. These results suggest that HyFunc offers a more efficient paradigm for agentic AI. Our code is publicly available at https://github.com/MrBlankness/HyFunc.
Bridging Global Intent with Local Details: A Hierarchical Representation Approach for Semantic Validation in Text-to-SQL
Dec 28, 2025Abstract:Text-to-SQL translates natural language questions into SQL statements grounded in a target database schema. Ensuring the reliability and executability of such systems requires validating generated SQL, but most existing approaches focus only on syntactic correctness, with few addressing semantic validation (detecting misalignments between questions and SQL). As a consequence, effective semantic validation still faces two key challenges: capturing both global user intent and SQL structural details, and constructing high-quality fine-grained sub-SQL annotations. To tackle these, we introduce HEROSQL, a hierarchical SQL representation approach that integrates global intent (via Logical Plans, LPs) and local details (via Abstract Syntax Trees, ASTs). To enable better information propagation, we employ a Nested Message Passing Neural Network (NMPNN) to capture inherent relational information in SQL and aggregate schema-guided semantics across LPs and ASTs. Additionally, to generate high-quality negative samples, we propose an AST-driven sub-SQL augmentation strategy, supporting robust optimization of fine-grained semantic inconsistencies. Extensive experiments conducted on Text-to-SQL validation benchmarks (both in-domain and out-of-domain settings) demonstrate that our approach outperforms existing state-of-the-art methods, achieving an average 9.40% improvement of AUPRC and 12.35% of AUROC in identifying semantic inconsistencies. It excels at detecting fine-grained semantic errors, provides large language models with more granular feedback, and ultimately enhances the reliability and interpretability of data querying platforms.
Magical: Medical Lay Language Generation via Semantic Invariance and Layperson-tailored Adaptation
Aug 12, 2025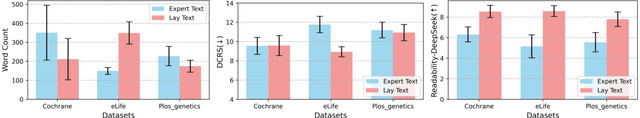
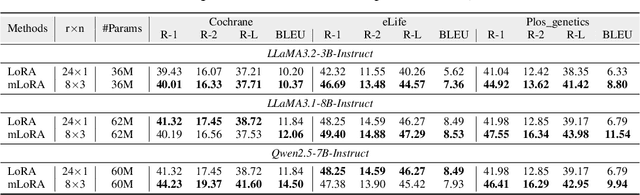
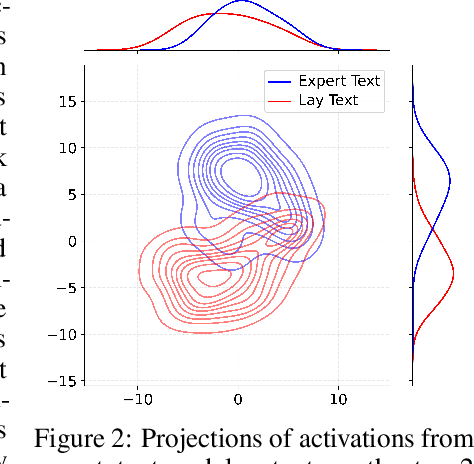
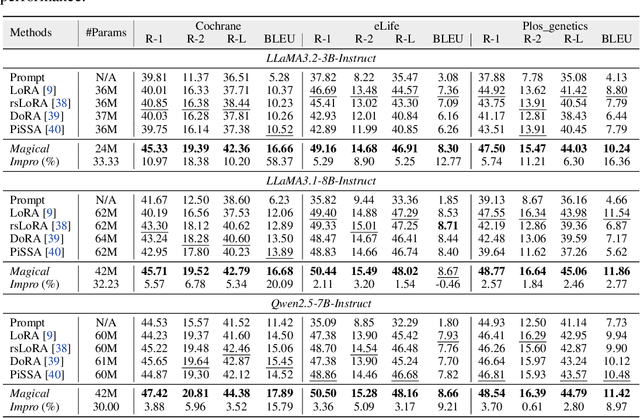
Abstract:Medical Lay Language Generation (MLLG) plays a vital role in improving the accessibility of complex scientific content for broader audiences. Recent literature to MLLG commonly employ parameter-efficient fine-tuning methods such as Low-Rank Adaptation (LoRA) to fine-tuning large language models (LLMs) using paired expert-lay language datasets. However, LoRA struggles with the challenges posed by multi-source heterogeneous MLLG datasets. Specifically, through a series of exploratory experiments, we reveal that standard LoRA fail to meet the requirement for semantic fidelity and diverse lay-style generation in MLLG task. To address these limitations, we propose Magical, an asymmetric LoRA architecture tailored for MLLG under heterogeneous data scenarios. Magical employs a shared matrix $A$ for abstractive summarization, along with multiple isolated matrices $B$ for diverse lay-style generation. To preserve semantic fidelity during the lay language generation process, Magical introduces a Semantic Invariance Constraint to mitigate semantic subspace shifts on matrix $A$. Furthermore, to better adapt to diverse lay-style generation, Magical incorporates the Recommendation-guided Switch, an externally interface to prompt the LLM to switch between different matrices $B$. Experimental results on three real-world lay language generation datasets demonstrate that Magical consistently outperforms prompt-based methods, vanilla LoRA, and its recent variants, while also reducing trainable parameters by 31.66%.
LearNAT: Learning NL2SQL with AST-guided Task Decomposition for Large Language Models
Apr 03, 2025Abstract:Natural Language to SQL (NL2SQL) has emerged as a critical task for enabling seamless interaction with databases. Recent advancements in Large Language Models (LLMs) have demonstrated remarkable performance in this domain. However, existing NL2SQL methods predominantly rely on closed-source LLMs leveraging prompt engineering, while open-source models typically require fine-tuning to acquire domain-specific knowledge. Despite these efforts, open-source LLMs struggle with complex NL2SQL tasks due to the indirect expression of user query objectives and the semantic gap between user queries and database schemas. Inspired by the application of reinforcement learning in mathematical problem-solving to encourage step-by-step reasoning in LLMs, we propose LearNAT (Learning NL2SQL with AST-guided Task Decomposition), a novel framework that improves the performance of open-source LLMs on complex NL2SQL tasks through task decomposition and reinforcement learning. LearNAT introduces three key components: (1) a Decomposition Synthesis Procedure that leverages Abstract Syntax Trees (ASTs) to guide efficient search and pruning strategies for task decomposition, (2) Margin-aware Reinforcement Learning, which employs fine-grained step-level optimization via DPO with AST margins, and (3) Adaptive Demonstration Reasoning, a mechanism for dynamically selecting relevant examples to enhance decomposition capabilities. Extensive experiments on two benchmark datasets, Spider and BIRD, demonstrate that LearNAT enables a 7B-parameter open-source LLM to achieve performance comparable to GPT-4, while offering improved efficiency and accessibility.
TPO: Aligning Large Language Models with Multi-branch & Multi-step Preference Trees
Oct 10, 2024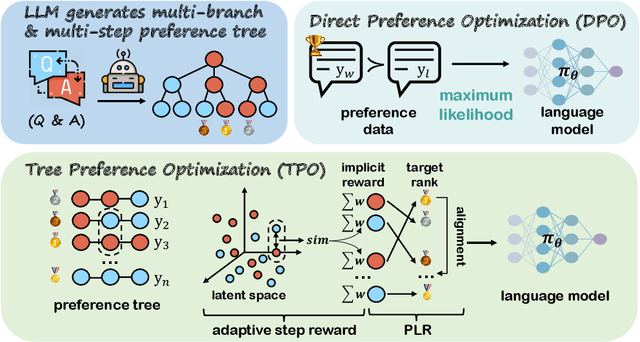
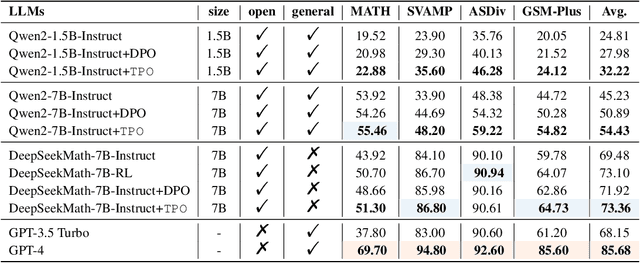

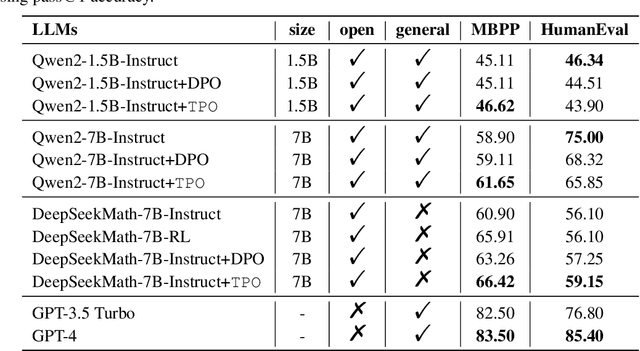
Abstract:In the domain of complex reasoning tasks, such as mathematical reasoning, recent advancements have proposed the use of Direct Preference Optimization (DPO) to suppress output of dispreferred responses, thereby enhancing the long-chain reasoning capabilities of large language models (LLMs). To this end, these studies employed LLMs to generate preference trees via Tree-of-thoughts (ToT) and sample the paired preference responses required by the DPO algorithm. However, the DPO algorithm based on binary preference optimization is unable to learn multiple responses with varying degrees of preference/dispreference that provided by the preference trees, resulting in incomplete preference learning. In this work, we introduce Tree Preference Optimization (TPO), that does not sample paired preference responses from the preference tree; instead, it directly learns from the entire preference tree during the fine-tuning. Specifically, TPO formulates the language model alignment as a Preference List Ranking problem, where the policy can potentially learn more effectively from a ranked preference list of responses given the prompt. In addition, to further assist LLMs in identifying discriminative steps within long-chain reasoning and increase the relative reward margin in the preference list, TPO utilizes Adaptive Step Reward to adjust the reward values of each step in trajectory for performing fine-grained preference optimization. We carry out extensive experiments on mathematical reasoning tasks to evaluate TPO. The experimental results indicate that TPO consistently outperforms DPO across three public large language models on four datasets.
RevGNN: Negative Sampling Enhanced Contrastive Graph Learning for Academic Reviewer Recommendation
Jul 30, 2024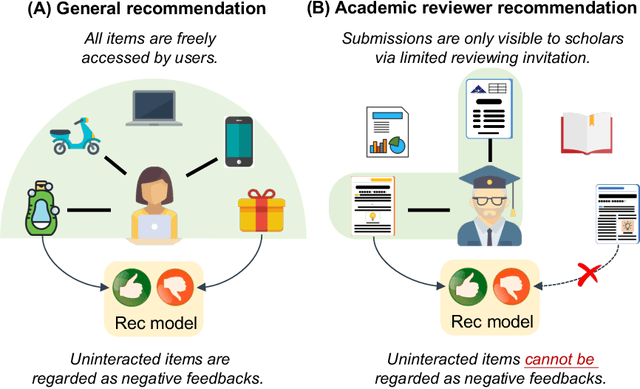
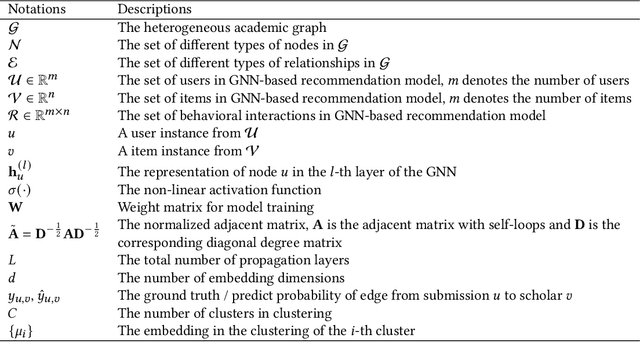
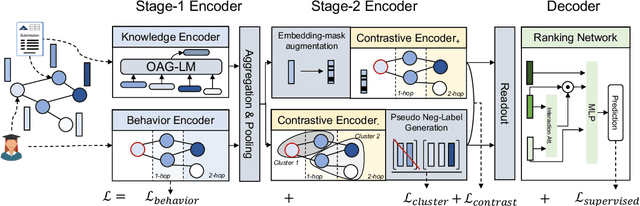
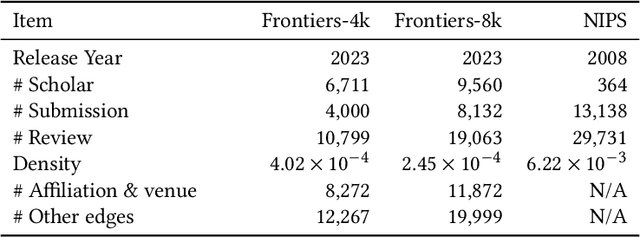
Abstract:Acquiring reviewers for academic submissions is a challenging recommendation scenario. Recent graph learning-driven models have made remarkable progress in the field of recommendation, but their performance in the academic reviewer recommendation task may suffer from a significant false negative issue. This arises from the assumption that unobserved edges represent negative samples. In fact, the mechanism of anonymous review results in inadequate exposure of interactions between reviewers and submissions, leading to a higher number of unobserved interactions compared to those caused by reviewers declining to participate. Therefore, investigating how to better comprehend the negative labeling of unobserved interactions in academic reviewer recommendations is a significant challenge. This study aims to tackle the ambiguous nature of unobserved interactions in academic reviewer recommendations. Specifically, we propose an unsupervised Pseudo Neg-Label strategy to enhance graph contrastive learning (GCL) for recommending reviewers for academic submissions, which we call RevGNN. RevGNN utilizes a two-stage encoder structure that encodes both scientific knowledge and behavior using Pseudo Neg-Label to approximate review preference. Extensive experiments on three real-world datasets demonstrate that RevGNN outperforms all baselines across four metrics. Additionally, detailed further analyses confirm the effectiveness of each component in RevGNN.
Is larger always better? Evaluating and prompting large language models for non-generative medical tasks
Jul 26, 2024Abstract:The use of Large Language Models (LLMs) in medicine is growing, but their ability to handle both structured Electronic Health Record (EHR) data and unstructured clinical notes is not well-studied. This study benchmarks various models, including GPT-based LLMs, BERT-based models, and traditional clinical predictive models, for non-generative medical tasks utilizing renowned datasets. We assessed 14 language models (9 GPT-based and 5 BERT-based) and 7 traditional predictive models using the MIMIC dataset (ICU patient records) and the TJH dataset (early COVID-19 EHR data), focusing on tasks such as mortality and readmission prediction, disease hierarchy reconstruction, and biomedical sentence matching, comparing both zero-shot and finetuned performance. Results indicated that LLMs exhibited robust zero-shot predictive capabilities on structured EHR data when using well-designed prompting strategies, frequently surpassing traditional models. However, for unstructured medical texts, LLMs did not outperform finetuned BERT models, which excelled in both supervised and unsupervised tasks. Consequently, while LLMs are effective for zero-shot learning on structured data, finetuned BERT models are more suitable for unstructured texts, underscoring the importance of selecting models based on specific task requirements and data characteristics to optimize the application of NLP technology in healthcare.
LightM-UNet: Mamba Assists in Lightweight UNet for Medical Image Segmentation
Mar 11, 2024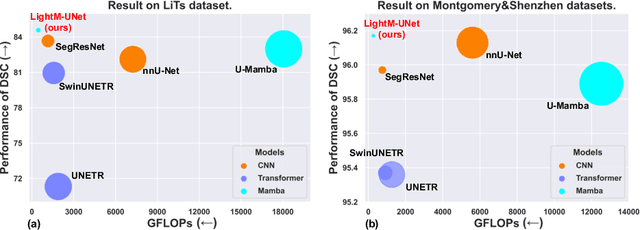
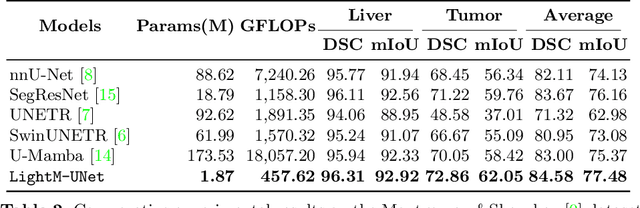
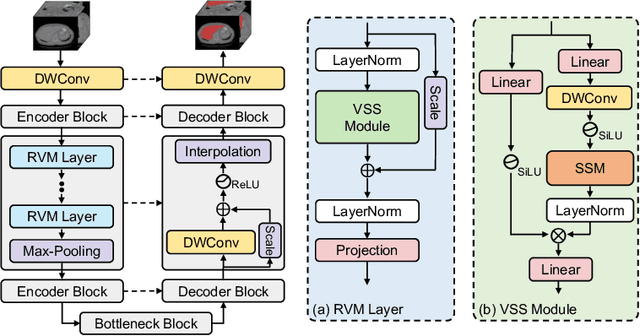
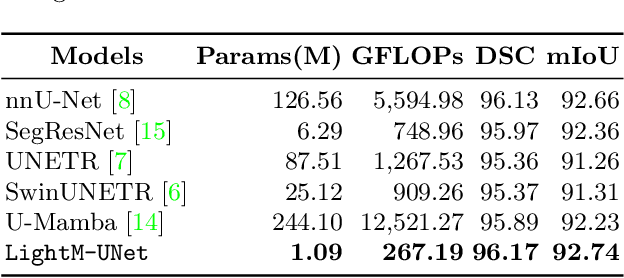
Abstract:UNet and its variants have been widely used in medical image segmentation. However, these models, especially those based on Transformer architectures, pose challenges due to their large number of parameters and computational loads, making them unsuitable for mobile health applications. Recently, State Space Models (SSMs), exemplified by Mamba, have emerged as competitive alternatives to CNN and Transformer architectures. Building upon this, we employ Mamba as a lightweight substitute for CNN and Transformer within UNet, aiming at tackling challenges stemming from computational resource limitations in real medical settings. To this end, we introduce the Lightweight Mamba UNet (LightM-UNet) that integrates Mamba and UNet in a lightweight framework. Specifically, LightM-UNet leverages the Residual Vision Mamba Layer in a pure Mamba fashion to extract deep semantic features and model long-range spatial dependencies, with linear computational complexity. Extensive experiments conducted on two real-world 2D/3D datasets demonstrate that LightM-UNet surpasses existing state-of-the-art literature. Notably, when compared to the renowned nnU-Net, LightM-UNet achieves superior segmentation performance while drastically reducing parameter and computation costs by 116x and 21x, respectively. This highlights the potential of Mamba in facilitating model lightweighting. Our code implementation is publicly available at https://github.com/MrBlankness/LightM-UNet.
Learnable Prompt as Pseudo-Imputation: Reassessing the Necessity of Traditional EHR Data Imputation in Downstream Clinical Prediction
Jan 30, 2024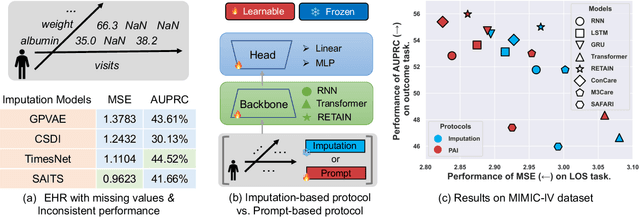
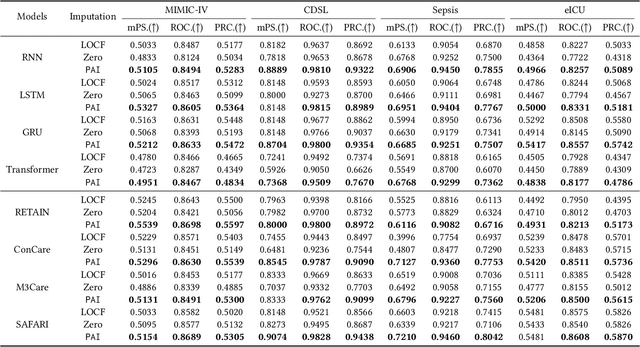
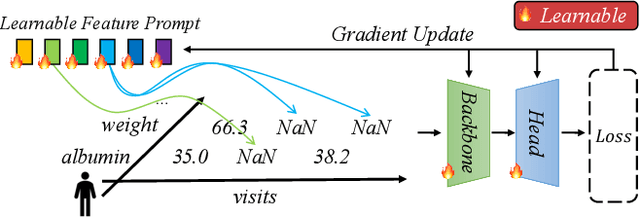
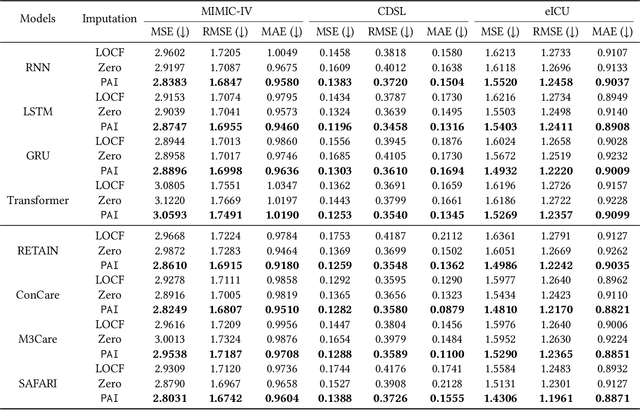
Abstract:Analyzing the health status of patients based on Electronic Health Records (EHR) is a fundamental research problem in medical informatics. The presence of extensive missing values in EHR makes it challenging for deep neural networks to directly model the patient's health status based on EHR. Existing deep learning training protocols require the use of statistical information or imputation models to reconstruct missing values; however, the protocols inject non-realistic data into downstream EHR analysis models, significantly limiting model performance. This paper introduces Learnable Prompt as Pseudo Imputation (PAI) as a new training protocol. PAI no longer introduces any imputed data but constructs a learnable prompt to model the implicit preferences of the downstream model for missing values, resulting in a significant performance improvement for all EHR analysis models. Additionally, our experiments show that PAI exhibits higher robustness in situations of data insufficiency and high missing rates. More importantly, in a real-world application involving cross-institutional data with zero-shot evaluation, PAI demonstrates stronger model generalization capabilities for non-overlapping features.
Prompting Large Language Models for Zero-Shot Clinical Prediction with Structured Longitudinal Electronic Health Record Data
Jan 25, 2024Abstract:The inherent complexity of structured longitudinal Electronic Health Records (EHR) data poses a significant challenge when integrated with Large Language Models (LLMs), which are traditionally tailored for natural language processing. Motivated by the urgent need for swift decision-making during new disease outbreaks, where traditional predictive models often fail due to a lack of historical data, this research investigates the adaptability of LLMs, like GPT-4, to EHR data. We particularly focus on their zero-shot capabilities, which enable them to make predictions in scenarios in which they haven't been explicitly trained. In response to the longitudinal, sparse, and knowledge-infused nature of EHR data, our prompting approach involves taking into account specific EHR characteristics such as units and reference ranges, and employing an in-context learning strategy that aligns with clinical contexts. Our comprehensive experiments on the MIMIC-IV and TJH datasets demonstrate that with our elaborately designed prompting framework, LLMs can improve prediction performance in key tasks such as mortality, length-of-stay, and 30-day readmission by about 35\%, surpassing ML models in few-shot settings. Our research underscores the potential of LLMs in enhancing clinical decision-making, especially in urgent healthcare situations like the outbreak of emerging diseases with no labeled data. The code is publicly available at https://github.com/yhzhu99/llm4healthcare for reproducibility.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge