Ewen M. Harrison
ColaCare: Enhancing Electronic Health Record Modeling through Large Language Model-Driven Multi-Agent Collaboration
Oct 03, 2024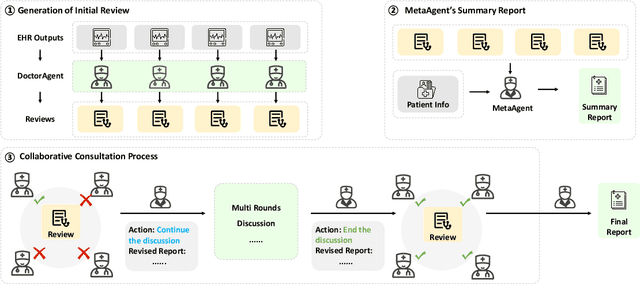
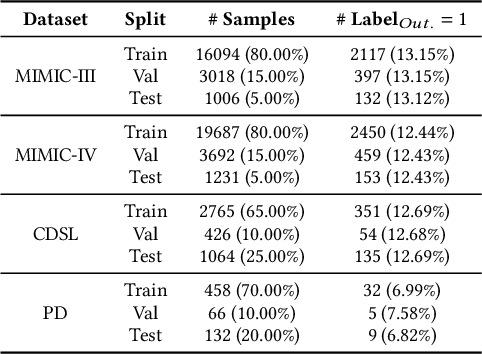
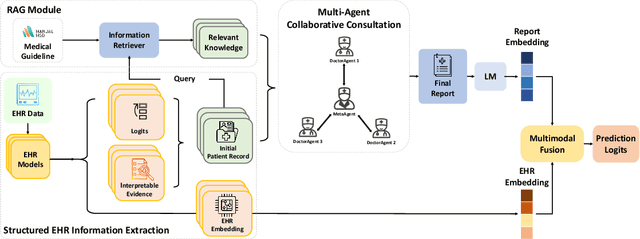
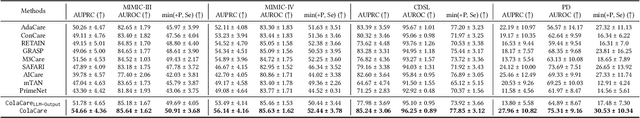
Abstract:We introduce ColaCare, a framework that enhances Electronic Health Record (EHR) modeling through multi-agent collaboration driven by Large Language Models (LLMs). Our approach seamlessly integrates domain-specific expert models with LLMs to bridge the gap between structured EHR data and text-based reasoning. Inspired by clinical consultations, ColaCare employs two types of agents: DoctorAgent and MetaAgent, which collaboratively analyze patient data. Expert models process and generate predictions from numerical EHR data, while LLM agents produce reasoning references and decision-making reports within the collaborative consultation framework. We additionally incorporate the Merck Manual of Diagnosis and Therapy (MSD) medical guideline within a retrieval-augmented generation (RAG) module for authoritative evidence support. Extensive experiments conducted on four distinct EHR datasets demonstrate ColaCare's superior performance in mortality prediction tasks, underscoring its potential to revolutionize clinical decision support systems and advance personalized precision medicine. The code, complete prompt templates, more case studies, etc. are publicly available at the anonymous link: https://colacare.netlify.app.
Is larger always better? Evaluating and prompting large language models for non-generative medical tasks
Jul 26, 2024Abstract:The use of Large Language Models (LLMs) in medicine is growing, but their ability to handle both structured Electronic Health Record (EHR) data and unstructured clinical notes is not well-studied. This study benchmarks various models, including GPT-based LLMs, BERT-based models, and traditional clinical predictive models, for non-generative medical tasks utilizing renowned datasets. We assessed 14 language models (9 GPT-based and 5 BERT-based) and 7 traditional predictive models using the MIMIC dataset (ICU patient records) and the TJH dataset (early COVID-19 EHR data), focusing on tasks such as mortality and readmission prediction, disease hierarchy reconstruction, and biomedical sentence matching, comparing both zero-shot and finetuned performance. Results indicated that LLMs exhibited robust zero-shot predictive capabilities on structured EHR data when using well-designed prompting strategies, frequently surpassing traditional models. However, for unstructured medical texts, LLMs did not outperform finetuned BERT models, which excelled in both supervised and unsupervised tasks. Consequently, while LLMs are effective for zero-shot learning on structured data, finetuned BERT models are more suitable for unstructured texts, underscoring the importance of selecting models based on specific task requirements and data characteristics to optimize the application of NLP technology in healthcare.
Prompting Large Language Models for Zero-Shot Clinical Prediction with Structured Longitudinal Electronic Health Record Data
Jan 25, 2024Abstract:The inherent complexity of structured longitudinal Electronic Health Records (EHR) data poses a significant challenge when integrated with Large Language Models (LLMs), which are traditionally tailored for natural language processing. Motivated by the urgent need for swift decision-making during new disease outbreaks, where traditional predictive models often fail due to a lack of historical data, this research investigates the adaptability of LLMs, like GPT-4, to EHR data. We particularly focus on their zero-shot capabilities, which enable them to make predictions in scenarios in which they haven't been explicitly trained. In response to the longitudinal, sparse, and knowledge-infused nature of EHR data, our prompting approach involves taking into account specific EHR characteristics such as units and reference ranges, and employing an in-context learning strategy that aligns with clinical contexts. Our comprehensive experiments on the MIMIC-IV and TJH datasets demonstrate that with our elaborately designed prompting framework, LLMs can improve prediction performance in key tasks such as mortality, length-of-stay, and 30-day readmission by about 35\%, surpassing ML models in few-shot settings. Our research underscores the potential of LLMs in enhancing clinical decision-making, especially in urgent healthcare situations like the outbreak of emerging diseases with no labeled data. The code is publicly available at https://github.com/yhzhu99/llm4healthcare for reproducibility.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge